Image classification
Rules for the classification of the images
The image classification will take place by generating a local database (a JSON
file called imagedb_cafos.json) for each observing night. For this purpose,
filabres will follow the rules provided in the instrument configuration
file configuration_cafos.yaml. Note that this file is embedded in the
distribution source code, under the directory filabres/instrument/.
Warning
Since the modification of the file configuration_cafos.yaml could change
the classification of the images perfomerd by filabres, in principle
this file should not be modified by a normal user. In any case, if you want
to change it, it will be necessary to reinstall the code for the changes to
be applied.
Warning
The following documentation has been prepared executing the commands in a terminal running the bash shell. If you are using the Z shell (that at the time of writing is the default in mac OS) it is important to remember that the wildcards employed in some filabres parameters need to be provided enclosed in double quotes.
In its first hierarchical level, this file configuration_cafos.yaml defines
the following keys: instname, version, requirements,
masterkeywords, and imagetypes:
instname: cafos
version: 2.3
requirements:
INSTRUME: 'CAFOS 2.2'
masterkeywords:
- NAXIS # number of data axes
- NAXIS1 # length of data axis 1
- NAXIS2 # length of data axis 2
- OBJECT # Target description
...
...
imagetypes:
bias:
...
...
flat-imaging:
...
...
flat-imaging-wollaston:
...
...
flat-spectroscopy:
...
...
arc:
...
...
science-imaging:
...
...
science-imaging-wollaston:
...
...
science-spectroscopy:
...
...
instname: the instrument name.version: the version of the instrument configuration file.requirements: the value of predefined FITS keywords that must be satisfied in order to classified the initial image within one of the categories defined below. If these requirements are not fulfilled, the image will be classified aswrong-instrument. For this particular example, a single requirement has been defined: the keywordINSTRUMEmust be defined in the FITS header and must be set toCAFOS 2.2.masterkeywords: list of keywords that must be stored in the local database.imagetypes: the different categories into which the initial images will be classified are provided as second-level dictionary of image types (e.g.:bias,flat-imaging,science-imaging, etc.). In order to proceed with this classification, each image type should be defined according to a third-level dictionary of keywords. For example, the image typebiasis defined as:imagetypes: bias: executable: True classification: calibration requirements: IMAGETYP: ['bias'] requirementx: EXPTIME: 0.0 QUANT975.LT.: 1000 signature: - CCDNAME - NAXIS1 - NAXIS2 - DATASEC - CCDBINX - CCDBINY maxtimespan_hours: 1
The corresponding section for
flat-imagingis:flat-imaging: executable: True classification: calibration requirements: IMAGETYP: ['flat', 'domeflat', 'twilight'] INSGRID: 'GRISM-11' INSPOFPI.NE.: 'Wollaston' requirementx: EXPTIME.GT.: 0 QUANT975.GT.: 5000 QUANT975.LT.: 60000 signature: - CCDNAME - NAXIS1 - NAXIS2 - DATASEC - CCDBINX - CCDBINY - INSGRID - INSFLID - INSPOFPI - INSPOROT maxtimespan_hours: 1 basicreduction: True
Another example: the
science-imagingtype is defined as:science-imaging: executable: True classification: science maxfieldview_arcmin: 16 requirements: IMAGETYP: ['science'] INSGRID: 'GRISM-11' INSPOFPI.NE.: 'Wollaston' requirementx: EXPTIME.GT.: 0 QUANT975.LT.: 60000 signature: - CCDNAME - NAXIS1 - NAXIS2 - DATASEC - CCDBINX - CCDBINY - INSGRID - INSFLID - INSPOFPI - INSPOROT - RA - DEC maxtimespan_hours: 0 basicreduction: True
The relevant keywords in this third-level dictionary are:
executable: if True, the reduction of this image type has been considered in filabres. Otherwise, the implementation of the reduction of this type of images is still pending.classification: so far only two possibilities are valid here:calibrationorscience. Note that calibration images will be combined to generate master calibrations (e.g., bias frames obtained in a predefined time span will be coadded), whereas science frames will be independently reduced (i.e., without image coaddition).requirements: conditions on FITS keywords that must be met in order to classify a particular image within the considered category. All these requirements are mandatory. Otherwise the image will be classified asunclassified. Important: note here thatIMAGETYPshould be defined as a list of possible names. For example, in the case of flats exposures in imaging mode, possible values for this keyword areflat,domeflatortwilight.requirementx: additional set of requirements . If any of these new requirements is not met, the image will be classified aswrong-<imagetype>(e.g.,bias-wrong,flat-imaging-wrong, etc.). Note that the separation of the requirements in these two sets (requirementsandrequirementx) allows to generate a classification where images initially classified within a given image type (because they verify all therequirements) can be flagged as suspicious because they exhibit an unexpected property (defined in therequirementx), such as anomalous exposure time or signal.maxtimespan_hours: calibration images with the same signature and within this maximum time span are combined prior to their reduction and generation of a single master calibration. Bias images are averaged using a median combination (note that the median combination gets rid of cosmic rays). Flat images are normalized prior to their median combination. Science images are reduced individually (i.e.,maxtimespan_hours: 0).basicreduction: logical flag that indicates whether science images are reduced using the master calibrations. This flag should always beTrue.
Note that images previously included in the file ignored_images.yaml will
be classified as ignored.
Inital image classification
The image classification is performed by starting the reduction step
initialize. Reduction steps are executed with the argument
-rs/--reduction_step.
Just to see the available reduction steps, execute:
$ filabres -rs
ERROR: missing reduction step / image type!
Initial options are:
- initialize (available: True)
- bias (available: True)
- flat-imaging (available: True)
- flat-imaging-wollaston (available: False)
- flat-spectroscopy (available: False)
- arc (available: False)
- science-imaging (available: True)
- science-imaging-wollaston (available: False)
- science-spectroscopy (available: False)
Next to each displayed reduction step you can see whether the considered reduction step is actually available or not (not available means that filabres will classify the corresponding images within that category but the reduction of those images is still missing in the code).
Since we are now interested in starting with the image classification, execute: (note that this can take several minutes!):
$ filabres -rs initialize
* Number of nights found: 58
* Working with night 170225_t2_CAFOS (1/58) ---> 140 FITS files
* Working with night 170226_t2_CAFOS (2/58) ---> 55 FITS files
...
...
* Working with night 171228_t2_CAFOS (57/58) ---> 50 FITS files
* Working with night 171230_t2_CAFOS (58/58) ---> 383 FITS files
* program STOP
A few warnings may be raised during the execution of the program. In particular
for the CAFOS 2017 data, the MJD-OBS is negative in some images and
filabres recomputes it. In other cases, HIERARCHCAHA DET CCDS is found,
when it sould be HIERARCH CAHA DET CCDS.
After the execution of previous command, a new subdirectory lists should
have appear in your working directory, containing subdirectories for all the
observing nights:
$ ls lists/
170225_t2_CAFOS/ 170524_t2_CAFOS/ 170807_t2_CAFOS/ 171108_t2_CAFOS/
170226_t2_CAFOS/ 170525_t2_CAFOS/ 170809_t2_CAFOS/ 171116_t2_CAFOS/
170319_t2_CAFOS/ 170526_t2_CAFOS/ 170811_t2_CAFOS/ 171120_t2_CAFOS/
170331_t2_CAFOS/ 170527_t2_CAFOS/ 170825_t2_CAFOS/ 171121_t2_CAFOS/
170403_t2_CAFOS/ 170528_t2_CAFOS/ 170903_t2_CAFOS/ 171209_t2_CAFOS/
170408_t2_CAFOS/ 170601_t2_CAFOS/ 170918_t2_CAFOS/ 171217_t2_CAFOS/
170420_t2_CAFOS/ 170602_t2_CAFOS/ 170926_t2_CAFOS/ 171218_t2_CAFOS/
170422_t2_CAFOS/ 170621_t2_CAFOS/ 170928_t2_CAFOS/ 171219_t2_CAFOS/
170502_t2_CAFOS/ 170627_t2_CAFOS/ 170929_t2_CAFOS/ 171221_t2_CAFOS/
170505_t2_CAFOS/ 170628_t2_CAFOS/ 171002_t2_CAFOS/ 171223_t2_CAFOS/
170506_t2_CAFOS/ 170629_t2_CAFOS/ 171008_t2_CAFOS/ 171225_t2_CAFOS/
170507_t2_CAFOS/ 170713_t2_CAFOS/ 171011_t2_CAFOS/ 171228_t2_CAFOS/
170517_t2_CAFOS/ 170720_t2_CAFOS/ 171015_t2_CAFOS/ 171230_t2_CAFOS/
170518_t2_CAFOS/ 170724_t2_CAFOS/ 171016_t2_CAFOS/
170519_t2_CAFOS/ 170731_t2_CAFOS/ 171101_t2_CAFOS/
Within each night, a file imagedb_cafos.json should have been created,
storing the image classification, as well as an additional file
imagedb_cafos.log storing a log with the result of the classification
(together with those WARNINGS raised during the image classification).
$ ls lists/170225_t2_CAFOS/
imagedb_cafos.json imagedb_cafos.log
All the warnings raised in the classification of the CAFOS 2017 data can safely be ignored.
Examine the image classification
Select image type
Although you can always try to open any of the files imagedb_cafos.json
directly (using a proper JSON editor), filabres provides an easier way to
examine the image classification previously performed (using the argument
-lc/--list_classified <imagetype>).
For example, to list the different image types available:
$ filabres -lc
Valid imagetypes:
- bias
- flat-imaging
- flat-imaging-wollaston
- flat-spectroscopy
- arc
- science-imaging
- science-imaging-wollaston
- science-spectroscopy
- wrong-bias
- wrong-flat-imaging
- wrong-flat-imaging-wollaston
- wrong-flat-spectroscopy
- wrong-arc
- wrong-science-imaging
- wrong-science-imaging-wollaston
- wrong-science-spectroscopy
- wrong-instrument
- ignored
- unclassified
You can repeat the same command by adding any of the above image types:
$ filabres -lc bias
file
1 /Volumes/NicoPassport/CAHA/CAFOS2017/170225_t2_CAFOS/caf-20170224-21:27:48-cal-krek.fits
2 /Volumes/NicoPassport/CAHA/CAFOS2017/170225_t2_CAFOS/caf-20170224-21:29:09-cal-krek.fits
3 /Volumes/NicoPassport/CAHA/CAFOS2017/170225_t2_CAFOS/caf-20170224-21:30:31-cal-krek.fits
...
...
824 /Volumes/NicoPassport/CAHA/CAFOS2017/171230_t2_CAFOS/caf-20171229-10:16:48-cal-lilj.fits
825 /Volumes/NicoPassport/CAHA/CAFOS2017/171230_t2_CAFOS/caf-20171229-10:17:24-cal-lilj.fits
826 /Volumes/NicoPassport/CAHA/CAFOS2017/171230_t2_CAFOS/caf-20171229-10:18:00-cal-lilj.fits
Total: 826 files
By default the list displays the full path to the original files.
Select image type and observing nights
It is possible to constraint the list of files to those corresponding to a
given subset of nights (using the argument -n/--night <night>; wildcards
are valid here):
$ filabres -lc bias -n 1702*
file
1 /Volumes/NicoPassport/CAHA/CAFOS2017/170225_t2_CAFOS/caf-20170224-21:27:48-cal-krek.fits
2 /Volumes/NicoPassport/CAHA/CAFOS2017/170225_t2_CAFOS/caf-20170224-21:29:09-cal-krek.fits
3 /Volumes/NicoPassport/CAHA/CAFOS2017/170225_t2_CAFOS/caf-20170224-21:30:31-cal-krek.fits
...
...
28 /Volumes/NicoPassport/CAHA/CAFOS2017/170226_t2_CAFOS/caf-20170226-11:47:59-cal-bomd.fits
29 /Volumes/NicoPassport/CAHA/CAFOS2017/170226_t2_CAFOS/caf-20170226-11:49:11-cal-bomd.fits
30 /Volumes/NicoPassport/CAHA/CAFOS2017/170226_t2_CAFOS/caf-20170226-11:50:23-cal-bomd.fits
Total: 30 files
Remember: when using wildcards in a terminal running the Z shell, you need to enclose the corresponding item with double quotes:
$ filabres -lc bias -n "1702*"
Select image type and relevant keywords
You can also display the values of relevant keywords belonging to the
masterkeywords list in the file configuration_cafos.yaml. If you don’t
remember them, don’t worry: use first -k/--keyword all to display all the
available keywords:
$ filabres -lc bias -k all
Valid keywords: ['NAXIS', 'NAXIS1', 'NAXIS2', 'OBJECT', 'RA', 'DEC',
'EQUINOX', 'DATE', 'MJD-OBS', 'AIRMASS', 'EXPTIME', 'INSTRUME', 'CCDNAME',
'ORIGSECX', 'ORIGSECY', 'CCDSEC', 'BIASSEC', 'DATASEC', 'CCDBINX',
'CCDBINY', 'IMAGETYP', 'INSTRMOD', 'INSAPID', 'INSTRSCL', 'INSTRPIX',
'INSTRPX0', 'INSTRPY0', 'INSFLID', 'INSFLNAM', 'INSGRID', 'INSGRNAM',
'INSGRROT', 'INSGRWL0', 'INSGRRES', 'INSPOFPI', 'INSPOROT', 'INSFPZ',
'INSFPWL', 'INSFPDWL', 'INSFPORD', 'INSCALST', 'INSCALID', 'INSCALNM',
'NPOINTS', 'FMINIMUM', 'QUANT025', 'QUANT159', 'QUANT250', 'QUANT500',
'QUANT750', 'QUANT841', 'QUANT975', 'FMAXIMUM', 'ROBUSTSTD']
Note
Note that, apart from the keywords belonging to the masterkeywords list
in the file configuration_cafos.yaml, some additional statistical
keywords are also available:
NPOINTS: number of points in the imageFMINIMUM,FMAXIMUM: mininum and maximum signal in the image.QUANT025,QUANT159,…QUANT975: 0.025, 0.159, 0.250, 0.500, 0.750, 0.841 and 0.975 quantiles of the data.ROBUSTSTD: robust estimate of the standard deviation, computed as 0.7413*(QUANT750-QUANT250).
Let’s display the values of a few of keywords: QUANT500 (the image median),
QUANT975 (the quantile 0.975 of the image), and ROBUSTSTD (the robust
standard deviation of the image):
$ filabres -lc bias -k quant500 -k quant975 -k robuststd
QUANT500 QUANT975 ROBUSTSTD file
1 666.00000 686.00000 11.11950 /Volumes/NicoPassport/CAHA/CAFOS2017/170225_t2_CAFOS/caf-20170224-21:27:48-cal-krek.fits
2 666.00000 687.00000 10.37820 /Volumes/NicoPassport/CAHA/CAFOS2017/170225_t2_CAFOS/caf-20170224-21:29:09-cal-krek.fits
3 666.00000 683.00000 10.37820 /Volumes/NicoPassport/CAHA/CAFOS2017/170225_t2_CAFOS/caf-20170224-21:30:31-cal-krek.fits
...
...
824 658.00000 680.00000 11.11950 /Volumes/NicoPassport/CAHA/CAFOS2017/171230_t2_CAFOS/caf-20171229-10:16:48-cal-lilj.fits
825 658.00000 680.00000 11.11950 /Volumes/NicoPassport/CAHA/CAFOS2017/171230_t2_CAFOS/caf-20171229-10:17:24-cal-lilj.fits
826 658.00000 680.00000 11.11950 /Volumes/NicoPassport/CAHA/CAFOS2017/171230_t2_CAFOS/caf-20171229-10:18:00-cal-lilj.fits
Total: 826 files
Note that each keyword is preceded by -k (following the astropy convention
for the fitsheader utility).
If instead of using -k/--keyword you use -ks/--keyword_sort, the list
will be sorted according to the selected keywords (several keys can be used for
a hierarchical sorting):
$ filabres -lc bias -k quant500 -k quant975 -ks robuststd
QUANT500 QUANT975 ROBUSTSTD file
456 661.40002 666.70001 2.81693 /Volumes/NicoPassport/CAHA/CAFOS2017/170929_t2_CAFOS/caf-20170929-13:52:35-cal-bias.fits
206 667.00000 683.00000 6.67170 /Volumes/NicoPassport/CAHA/CAFOS2017/170526_t2_CAFOS/caf-20170526-15:44:34-cal-boeh.fits
207 667.00000 683.00000 6.67170 /Volumes/NicoPassport/CAHA/CAFOS2017/170526_t2_CAFOS/caf-20170526-15:45:45-cal-boeh.fits
...
...
241 723.00000 776.00000 25.94550 /Volumes/NicoPassport/CAHA/CAFOS2017/170601_t2_CAFOS/caf-20170601-13:12:14-cal-bomd.fits
245 723.00000 776.00000 25.94550 /Volumes/NicoPassport/CAHA/CAFOS2017/170601_t2_CAFOS/caf-20170601-13:17:01-cal-bomd.fits
311 693.00000 729.00000 25.94550 /Volumes/NicoPassport/CAHA/CAFOS2017/170628_t2_CAFOS/caf-20170628-17:29:10-cal-pelm.fits
Total: 826 files
Now the values in the column ROBUSTSTD have appeared sorted
(note that the sorted keys are always the last columns preceding the file
column in the table, independently of their location in the command line).
Is is also possible to generate plots with the selected keywords. For that
purpose, employ the -pxy argument:
$ filabres -lc bias -k mjd-obs -k quant500 -k quant975 -ks robuststd -pxy
...
...
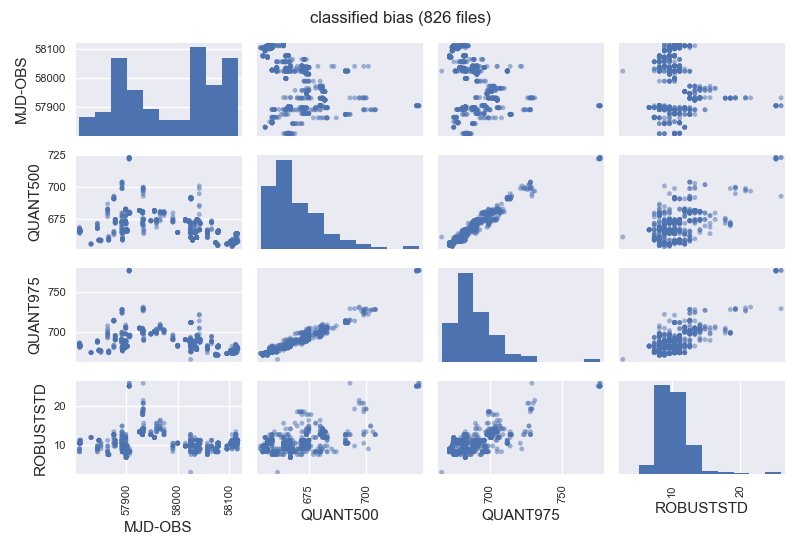
The previous image is a pairs plot, that allows to see both the distribution of values of each (numeric) keyword and the relationships between any two of them.
Filtering the list
It is also possible to filter the displayed list by using --filter
EXPRESSION, where EXPRESSION is any generic logical expression involving
valid keywords (i.e., belonging to the masterkeywords list) written as a
valid Python command, but using the special syntax k[<keyword>] to specify
the keywords. Note that arithmetic and logical operators are valid in
EXPRESSION.
For example, we can filter imposing fixed image dimensions:
$ filabres -lc bias -k naxis1 -k naxis2 --filter "k[naxis1]==1650 and k[naxis2]==1650"
NAXIS1 NAXIS2 file
1 1650 1650 /Volumes/NicoPassport/CAHA/CAFOS2017/170225_t2_CAFOS/caf-20170224-21:27:48-cal-krek.fits
2 1650 1650 /Volumes/NicoPassport/CAHA/CAFOS2017/170225_t2_CAFOS/caf-20170224-21:29:09-cal-krek.fits
3 1650 1650 /Volumes/NicoPassport/CAHA/CAFOS2017/170225_t2_CAFOS/caf-20170224-21:30:31-cal-krek.fits
4 1650 1650 /Volumes/NicoPassport/CAHA/CAFOS2017/170225_t2_CAFOS/caf-20170224-21:31:52-cal-krek.fits
5 1650 1650 /Volumes/NicoPassport/CAHA/CAFOS2017/170225_t2_CAFOS/caf-20170224-21:33:14-cal-krek.fits
6 1650 1650 /Volumes/NicoPassport/CAHA/CAFOS2017/170225_t2_CAFOS/caf-20170224-21:34:36-cal-krek.fits
7 1650 1650 /Volumes/NicoPassport/CAHA/CAFOS2017/170225_t2_CAFOS/caf-20170224-21:35:57-cal-krek.fits
8 1650 1650 /Volumes/NicoPassport/CAHA/CAFOS2017/170225_t2_CAFOS/caf-20170224-21:37:19-cal-krek.fits
9 1650 1650 /Volumes/NicoPassport/CAHA/CAFOS2017/170225_t2_CAFOS/caf-20170224-21:38:41-cal-krek.fits
10 1650 1650 /Volumes/NicoPassport/CAHA/CAFOS2017/170225_t2_CAFOS/caf-20170224-21:40:03-cal-krek.fits
Total: 10 files
The keywords employed in EXPRESSION do not have to be explicitly shown in
the table using -k <keyword> (in the last example they appear because we
want to check that the --filter argument is working properly).
Is there something wrong with the image classification?
Before moving to the reduction of the calibration images, it is important to check the image classification. In this sense, a few image types should be revised, as shown in the following subsections.
Note
We are going to skip the image types wrong-flat-spectroscopy,
wrong-arc and wrong-science-spectroscopy because we are initially
interested in working only with direct imaging calibrations and
observations.
Wrong instrument
There should be no images classified as wrong-instrument:
$ filabres -lc wrong-instrument
Total: 0 files
Unclassified
These are images that could not be classified according to the rules
defined in configuration_cafos.yaml:
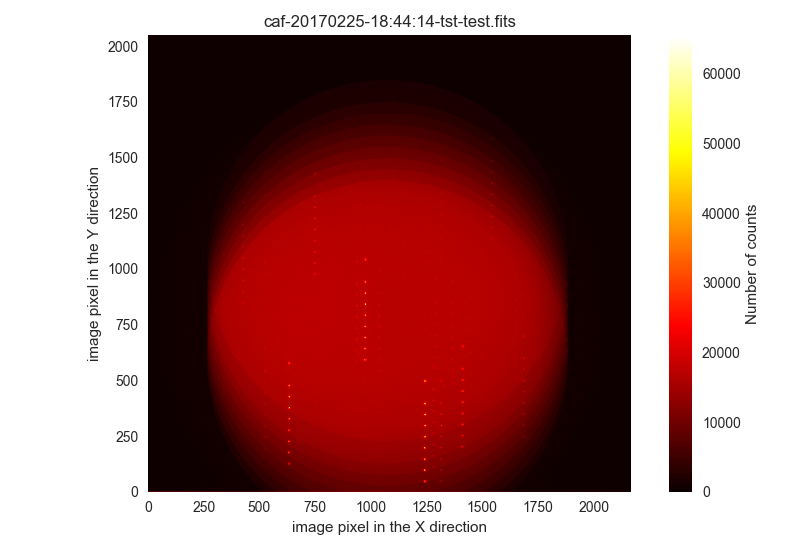
$ filabres -lc unclassified -k object
OBJECT file
1 [focus] Telescope /Volumes/NicoPassport/CAHA/CAFOS2017/170225_t2_CAFOS/caf-20170225-18:44:14-tst-test.fits
2 [focus] Telescope /Volumes/NicoPassport/CAHA/CAFOS2017/170505_t2_CAFOS/caf-20170506-02:53:44-tst-test.fits
3 ETALON_calibration /Volumes/NicoPassport/CAHA/CAFOS2017/170601_t2_CAFOS/caf-20170601-14:00:42-sci-etac.fits
4 ETALON_calibration /Volumes/NicoPassport/CAHA/CAFOS2017/170628_t2_CAFOS/caf-20170628-16:26:53-sci-etac.fits
5 ETALON_calibration /Volumes/NicoPassport/CAHA/CAFOS2017/170628_t2_CAFOS/caf-20170628-16:35:52-sci-etac.fits
6 [focus] Telescope /Volumes/NicoPassport/CAHA/CAFOS2017/170807_t2_CAFOS/caf-20170807-21:10:39-cal-schn.fits
Total: 6 files
Only 6 images appear in this category. The keyword OBJECT indicates that
they correspond to some auxiliary (telescope focus) or calibration (etalon)
images that we are not going to use in the reduction of our data. So, we can
safely ignore them.
Interstingly, filabres allows you to display all of them in sequence adding
the argument -pi (plot image):
$ filabres -lc unclassified -k object -pi
...
...
Here we show the first two images:
Ignored
These are simply the images that we have included in the file
ignored_images.yaml:
$ filabres -lc ignored
file
1 /Volumes/NicoPassport/CAHA/CAFOS2017/170506_t2_CAFOS/caf-20170505-09:57:54-cal-agui.fits
2 /Volumes/NicoPassport/CAHA/CAFOS2017/170506_t2_CAFOS/caf-20170505-09:58:58-cal-agui.fits
3 /Volumes/NicoPassport/CAHA/CAFOS2017/170506_t2_CAFOS/caf-20170505-10:00:02-cal-agui.fits
...
...
253 /Volumes/NicoPassport/CAHA/CAFOS2017/171219_t2_CAFOS/caf-20171218-17:43:07-cal-bard.fits
254 /Volumes/NicoPassport/CAHA/CAFOS2017/171219_t2_CAFOS/caf-20171218-17:45:23-cal-bard.fits
255 /Volumes/NicoPassport/CAHA/CAFOS2017/171219_t2_CAFOS/caf-20171218-17:47:46-cal-bard.fits
Total: 255 files
Wrong bias
These are the bias images that passed the requirements set in
configuration_cafos.yaml but not the requirmentx:
$ filabres -lc wrong-bias
Total: 0 files
None.
Wrong flat-imaging
These are the flat-imaging images that passed the requirements set in
configuration_cafos.yaml but not the requirmentx:
$ filabres -lc wrong-flat-imaging
...
...
Total: 228 files
There are 228 images in classified in this category. It is useful to check a few keywords:
$ filabres -lc wrong-flat-imaging --filter "k[exptime] == 0"
Total: 0 files
Nothing wrong with EXPTIME.
Let’s examine the signal, using the median QUANT500 and the quantile 0.975
(sorting in the latter):
$ filabres -lc wrong-flat-imaging -k quant500 -ks quant975
...
...
Total: 228 files
The last table reveals several cases:
Images with unexpectedly low signal:
$ filabres -lc wrong-flat-imaging -k quant500 -ks quant975 --filter "k[quant500] < 100" QUANT500 QUANT975 file 1 -0.01052 -0.00291 /Volumes/NicoPassport/CAHA/CAFOS2017/170929_t2_CAFOS/caf-20170929-14:38:37-cal-delt.fits 2 0.99903 1.00470 /Volumes/NicoPassport/CAHA/CAFOS2017/170929_t2_CAFOS/caf-20170929-14:40:39-cal-quot.fits Total: 2 files
There are only two images in this situation. The visual examination reveals that they lack even the bias level (the median
QUANT500should be around the usual bias signal, in this case slightly below 700 ADUs, which is not the case):$ filabres -lc wrong-flat-imaging -k quant500 -ks quant975 --filter "k[quant500] < 100" -pi ... ...
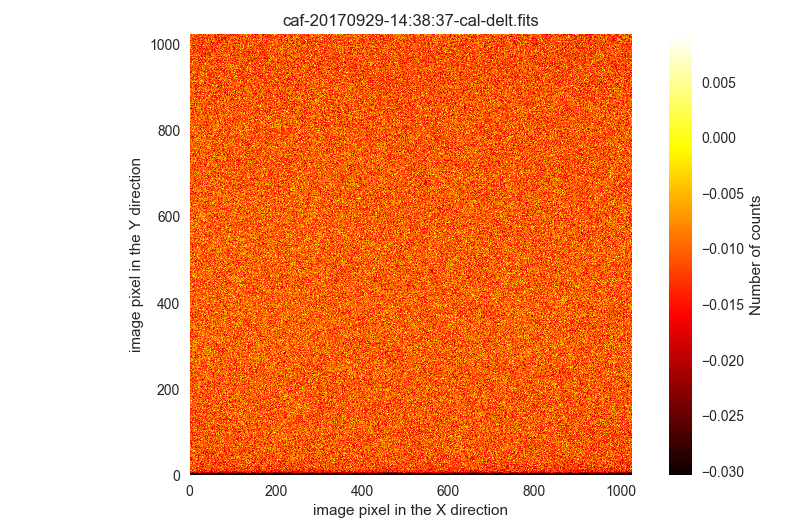
These two images will be ignored.
Images incompatible with being flat images attending to the signal level:
$ filabres -lc wrong-flat-imaging -k quant500 -ks quant975 -k imagetyp \ -k object -k exptime --filter "100 < k[quant500] < 800" QUANT500 IMAGETYP OBJECT EXPTIME QUANT975 file 5 705.0 flat [flat] sky R 0.5 731.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170929_t2_CAFOS/caf-20170929-18:27:21-cal-wenj.fits 3 706.0 flat [flat] sky R 1.0 732.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170929_t2_CAFOS/caf-20170929-18:23:42-cal-wenj.fits 4 706.0 flat [flat] sky R 0.5 732.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170929_t2_CAFOS/caf-20170929-18:25:39-cal-wenj.fits 6 705.0 flat [flat] sky R 10.0 732.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170929_t2_CAFOS/caf-20170929-18:28:45-cal-wenj.fits 1 706.0 flat [flat] sky R 0.5 733.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170929_t2_CAFOS/caf-20170929-18:21:50-cal-wenj.fits 2 706.0 flat [flat] sky R 15.0 733.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170929_t2_CAFOS/caf-20170929-18:22:37-cal-wenj.fits Total: 6 files
Note that although the exposure times are larger than zero, the signal is basically compatible with that expected in a bias frame. The visual inspection of these 6 images shows that this is the case:
$ filabres -lc wrong-flat-imaging -k quant500 -ks quant975 -k imagetyp \ -k object -k exptime --filter "100 < k[quant500] < 800" -pi ... ...
These 6 images are going to be ignored.
Flat images with low signal: these images can be selected by filtering those with a median value in the interval [800, 50000], and by looking for the word
flatin the description provided by the FITS keywordOBJECT(note here the flexibility of the--filterargument that allows to write an explicit Python logical expression):$ filabres -lc wrong-flat-imaging -k quant500 -ks quant975 -k object \ -k exptime --filter "800 < k[quant500] < 50000 and 'flat' in k[object].lower()" QUANT500 OBJECT EXPTIME QUANT975 file 2 1050.0 [Flat]Sky U 5.0 1096.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170628_t2_CAFOS/caf-20170628-20:17:39-cal-mirl.fits 1 3539.0 [Flat]Sky U 5.0 3696.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170627_t2_CAFOS/caf-20170627-20:07:30-cal-mirl.fits 3 3786.0 [Flat]Sky U 60.0 3947.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170628_t2_CAFOS/caf-20170628-20:18:47-cal-mirl.fits 4 4752.0 [Flat]Sky U 180.0 4952.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170628_t2_CAFOS/caf-20170628-20:23:30-cal-mirl.fits Total: 4 files
These images were classified as
wrong-flat-imagingbecauseQUANT975was lower than 5000 ADUs (this is one of therequirementxfor this type of images).We are going to modify the automatic image classification performed by filabres by forcing these 4 images to be reclassified as valid
flat-imagingframes. To perform this manual classification, the first step is the inclusion of those files in the file configuration YAML fileforced_classifications.yaml:1# file forced_classifications.yaml 2# 3# generated automatically by filabres v.1.3.0 4# 5# creation date 2023-03-10T18:19:57.374064 6# 7 8# sky flat with low signal 9night: 170627_t2_CAFOS 10enabled: True 11files: 12 - caf-20170627-20:07:30-cal-mirl.fits 13classification: flat-imaging 14 15--- 16 17# sky flat with low signal 18night: 170628_t2_CAFOS 19enabled: True 20files: 21 - caf-20170628-20:17:39-cal-mirl.fits 22 - caf-20170628-20:18:47-cal-mirl.fits 23 - caf-20170628-20:23:30-cal-mirl.fits 24classification: flat-imaging
Ignoring the initial comment lines (starting by
#), here we have created two blocks, separated by---(the YAML block separator). Important: the separator must not appear before the first block nor after the last block. Within each block, the following arguments must be provided:night: observing night. Note that wildcards can not be used here, although the same night label can appear in different blocks.enabled: this key indicates that the current block must be used (this keyword allows a quick way to disable a whole block by setting this value toFalse, without the need of removing it from this file).files: is the list of files to be considered within the specified night. The list of files can be provided by given the name of each file in separate line, preceded by an indented - symbol. Wildcards are valid here.classification: classification label for the images listed in the block.
Additional comment lines (starting by
#) or blank lines can be used in this YAML file.Once we have inserted the 4 problematic images within this file, we can repeat the classification of these files. Since the images correspond to two different nights, it is necessary to repeat the classification in the corresponding nights:
$ filabres -rs initialize -n 170627* --force ... ... $ filabres -rs initialize -n 170628* --force ... ...
Note that the argument
--forcehas been used to force the repetition of the reduction for those nights. Without this argument, filabres avoids the repetion because it detects that there is already a fileimagedb_cafos.jsonin the corresponding subdirectory night. By forcing the repetition we command filabres to override that file.Finally, it is possible to double check that the 4 images do not longer appear as classified as
wrong-flat-imaging:$ filabres -lc wrong-flat-imaging -k quant500 -ks quant975 -k object \ -k exptime --filter "800 < k[quant500] < 50000 and 'flat' in k[object].lower()" Total: 0 files
Saturated images: in this case we list images classified as
wrong-flat-imagingbut with aQUANT975above 60000 ADUs:$ filabres -lc wrong-flat-imaging -k exptime -k quant500 -k quant975 -k object \ --filter "k[quant975] >= 60000" EXPTIME QUANT500 QUANT975 OBJECT file 1 45.0 51930.0 60050.0 [Skyflat] r' /Volumes/NicoPassport/CAHA/CAFOS2017/170225_t2_CAFOS/caf-20170226-06:11:17-cal-krek.fits 2 2.0 57062.0 64891.0 [Skyflat] g' /Volumes/NicoPassport/CAHA/CAFOS2017/170225_t2_CAFOS/caf-20170226-06:22:28-cal-krek.fits 3 0.1 65531.0 65531.0 [flat] sky B /Volumes/NicoPassport/CAHA/CAFOS2017/170929_t2_CAFOS/caf-20170929-18:05:56-cal-wenj.fits 4 0.1 65531.0 65531.0 [flat] sky R /Volumes/NicoPassport/CAHA/CAFOS2017/170929_t2_CAFOS/caf-20170929-18:06:46-cal-wenj.fits 5 0.1 65531.0 65531.0 [flat] sky R /Volumes/NicoPassport/CAHA/CAFOS2017/170929_t2_CAFOS/caf-20170929-18:11:14-cal-wenj.fits 6 0.1 60313.0 61806.0 [flat] sky R /Volumes/NicoPassport/CAHA/CAFOS2017/170929_t2_CAFOS/caf-20170929-18:12:02-cal-wenj.fits Total: 6 files
These images are clearly saturated. You can display them using
-pi:$ filabres -lc wrong-flat-imaging -k exptime -k quant500 -k quant975 -k object \ --filter "k[quant975] >= 60000" -pi ... ...
The first two images are similar to the following:
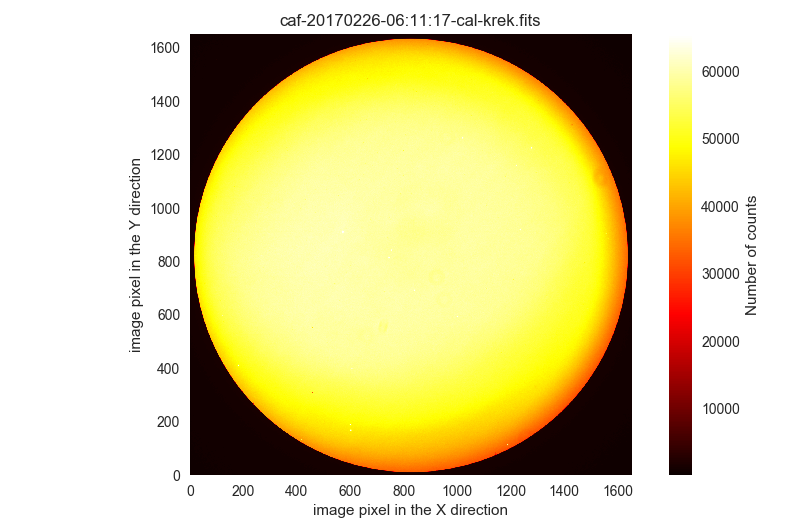
The next three images are similar to the following:
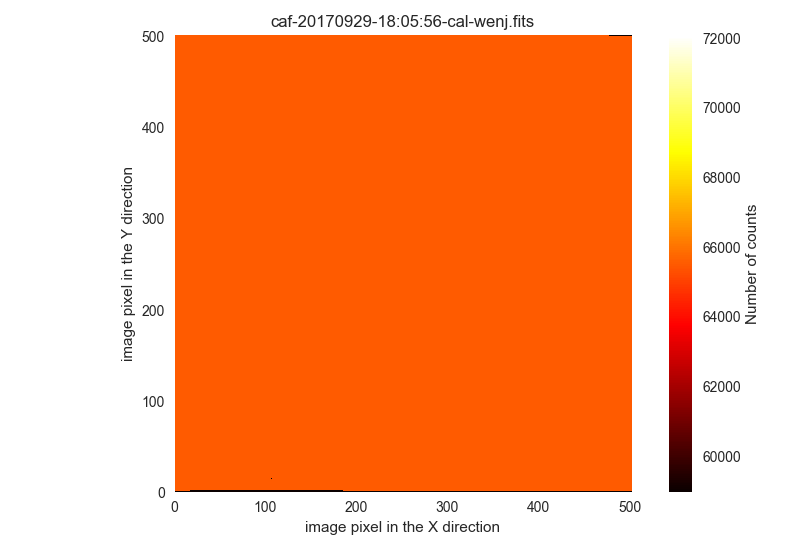
The last image is only saturated in some regions:
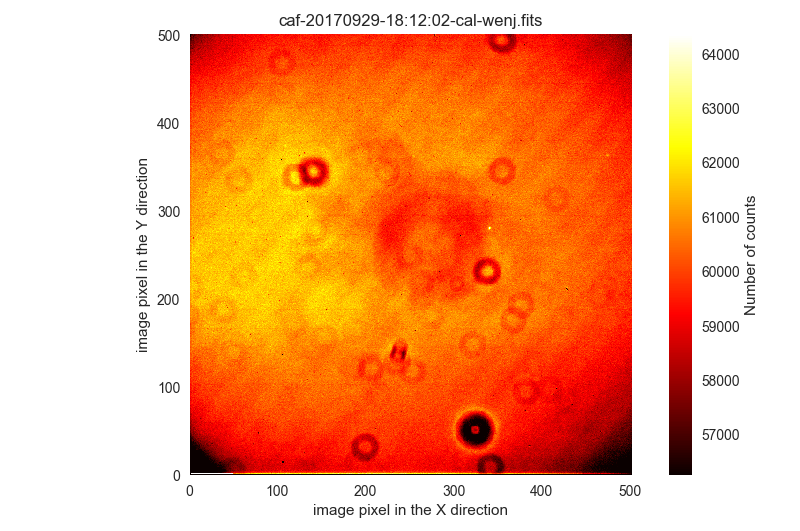
These six images will be ignored.
The remaining 210 files correspond to science images with erroneous
IMAGTYPset to flat:$ filabres -lc wrong-flat-imaging -k exptime -k quant500 -k quant975 -k object \ --filter "800 < k[quant500] < 50000" EXPTIME QUANT500 QUANT975 OBJECT file 1 60.0 898.0 946.0 HAT-P-12b B 60s /Volumes/NicoPassport/CAHA/CAFOS2017/170518_t2_CAFOS/caf-20170518-22:21:06-cal-alex.fits 2 60.0 898.0 946.0 HAT-P-12b B 60s /Volumes/NicoPassport/CAHA/CAFOS2017/170518_t2_CAFOS/caf-20170518-22:22:33-cal-alex.fits 3 60.0 898.0 946.0 HAT-P-12b B 60s /Volumes/NicoPassport/CAHA/CAFOS2017/170518_t2_CAFOS/caf-20170518-22:23:59-cal-alex.fits ... ... Total: 210 filesThe
OBJECTvalue in these images seems to indicate that they do not constitute calibration frames. These images correspond to two nights:170518and170601. Actually, the first 175 files seem to be science images of the same sky region (includeRAandDECin the table) obtained in170518:$ filabres -lc wrong-flat-imaging -k exptime -k quant500 -k quant975 -k object \ -k ra -k dec -n 170518* EXPTIME QUANT500 QUANT975 OBJECT RA DEC file 1 60.0 898.0 946.0 HAT-P-12b B 60s 209.50799 43.39571 /Volumes/NicoPassport/CAHA/CAFOS2017/170518_t2_CAFOS/caf-20170518-22:21:06-cal-alex.fits 2 60.0 898.0 946.0 HAT-P-12b B 60s 209.50799 43.39571 /Volumes/NicoPassport/CAHA/CAFOS2017/170518_t2_CAFOS/caf-20170518-22:22:33-cal-alex.fits 3 60.0 898.0 946.0 HAT-P-12b B 60s 209.50798 43.39572 /Volumes/NicoPassport/CAHA/CAFOS2017/170518_t2_CAFOS/caf-20170518-22:23:59-cal-alex.fits ... ... 173 60.0 1415.0 1499.0 HAT-P-12b B 60s 209.50657 43.39647 /Volumes/NicoPassport/CAHA/CAFOS2017/170518_t2_CAFOS/caf-20170519-02:31:34-cal-alex.fits 174 60.0 1419.0 1506.0 HAT-P-12b B 60s 209.50654 43.39652 /Volumes/NicoPassport/CAHA/CAFOS2017/170518_t2_CAFOS/caf-20170519-02:33:00-cal-alex.fits 175 60.0 1425.0 1512.0 HAT-P-12b B 60s 209.50654 43.39658 /Volumes/NicoPassport/CAHA/CAFOS2017/170518_t2_CAFOS/caf-20170519-02:34:26-cal-alex.fits Total: 175 filesThe (patient) visual inspection of these images show:
$ filabres -lc wrong-flat-imaging -k exptime -k quant500 -k quant975 -k object \ -k ra -k dec -n 170518* -pi ... ...
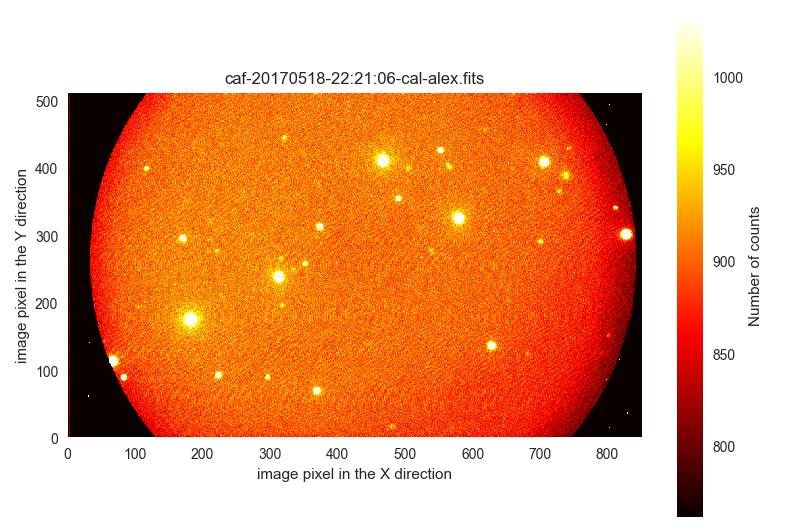
The 35 images from
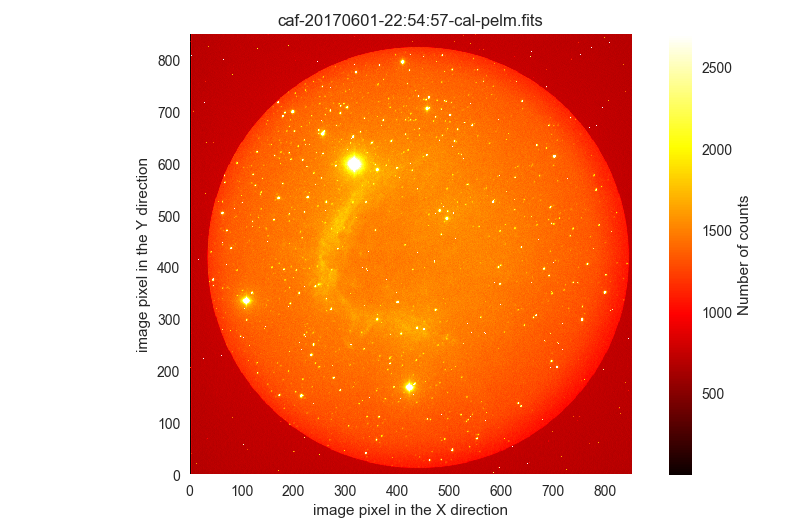
170601are also likely science frames:$ filabres -lc wrong-flat-imaging -k exptime -k quant500 -k quant975 -k object \ -k ra -k dec -n 170601* EXPTIME QUANT500 QUANT975 OBJECT RA DEC file 1 600.0 1354.0 1664.0 IC1396N ETALON+674/18 325.31573 58.37090 /Volumes/NicoPassport/CAHA/CAFOS2017/170601_t2_CAFOS/caf-20170601-22:54:57-cal-pelm.fits 2 600.0 1342.0 1648.0 IC1396N ETALON+674/18 325.32135 58.37090 /Volumes/NicoPassport/CAHA/CAFOS2017/170601_t2_CAFOS/caf-20170601-23:06:07-cal-pelm.fits 3 600.0 1330.0 1622.0 IC1396N ETALON+674/18 325.32152 58.37635 /Volumes/NicoPassport/CAHA/CAFOS2017/170601_t2_CAFOS/caf-20170601-23:16:42-cal-pelm.fits ... ... 33 30.0 1771.0 2105.0 IC1396W_2 R 323.69004 57.58110 /Volumes/NicoPassport/CAHA/CAFOS2017/170601_t2_CAFOS/caf-20170602-03:02:56-cal-pelm.fits 34 30.0 1771.0 2105.0 IC1396W_2 R 323.68997 57.58110 /Volumes/NicoPassport/CAHA/CAFOS2017/170601_t2_CAFOS/caf-20170602-03:04:00-cal-pelm.fits 35 30.0 1769.0 2107.0 IC1396W_2 R 323.68998 57.58111 /Volumes/NicoPassport/CAHA/CAFOS2017/170601_t2_CAFOS/caf-20170602-03:05:05-cal-pelm.fits Total: 35 filesThe visual inspection again reveals that this image set corresponds to science frames:
$ filabres -lc wrong-flat-imaging -k exptime -k quant500 -k quant975 -k object \ -k ra -k dec -n 170601* -pi ... ...
The easiest way to recover these 210 files erroneously classified as
wrong-flat-imaging, and that we have deduced should be classified asscience-imaging, is to modify the original FITS keywordIMAGETYP, initially set toflat, and reset it toscience. This can be easily done by inserting the corresponding files in the configuration YAML fileimage_header_corrections.yaml, as will be explained below. Note that it would also be possible to force the classification of those images asscience-imagingusing the configuration YAML fileforced_classifications.yaml, but the new approach seems more appropiate because it will also fix the wrong value of the FITS keywordIMAGETYPEand, in addition, filabres will check the correspondingrequirementsandrequirementxforscience-imaging.Since the number of files to be inserted in
image_header_corrections.yamlis large (175 for night170518and 35 for night170601), we are using a command line to generate a list of files suitable for cut and paste.For night
170518:$ filabres -lc wrong-flat-imaging -n 170518* -lm basic - caf-20170518-22:21:06-cal-alex.fits - caf-20170518-22:22:33-cal-alex.fits - caf-20170518-22:23:59-cal-alex.fits ... ... - caf-20170519-02:31:34-cal-alex.fits - caf-20170519-02:33:00-cal-alex.fits - caf-20170519-02:34:26-cal-alex.fits Total: 175 files
For night
170601:$ filabres -lc wrong-flat-imaging -n 170601* -lm basic - caf-20170601-22:54:57-cal-pelm.fits - caf-20170601-23:06:07-cal-pelm.fits - caf-20170601-23:16:42-cal-pelm.fits ... ... - caf-20170602-03:02:56-cal-pelm.fits - caf-20170602-03:04:00-cal-pelm.fits - caf-20170602-03:05:05-cal-pelm.fits Total: 35 files
These last two list of files starting by
-can be easily cut and pasted into theimage_header_corrections.yaml, so the contents of this file are:1# file image_header_corrections.yaml 2# 3# generated automatically by filabres v.1.3.0 4# 5# creation date 2023-03-10T18:19:57.373984 6# 7 8# erroneous wrong-flat-imaging: they are actually science exposures 9night: 170518_t2_CAFOS 10enabled: True 11files: 12 - caf-20170518-22:21:06-cal-alex.fits 13 - caf-20170518-22:22:33-cal-alex.fits 14 - caf-20170518-22:23:59-cal-alex.fits 15 - caf-20170518-22:26:08-cal-alex.fits 16 - caf-20170518-22:28:43-cal-alex.fits 17 - caf-20170518-22:30:10-cal-alex.fits 18 - caf-20170518-22:31:36-cal-alex.fits 19 - caf-20170518-22:33:03-cal-alex.fits 20 - caf-20170518-22:34:29-cal-alex.fits 21 - caf-20170518-22:35:55-cal-alex.fits 22 - caf-20170518-22:37:21-cal-alex.fits 23 - caf-20170518-22:38:48-cal-alex.fits 24 - caf-20170518-22:40:14-cal-alex.fits 25 - caf-20170518-22:41:40-cal-alex.fits 26 - caf-20170518-22:43:06-cal-alex.fits 27 - caf-20170518-22:44:32-cal-alex.fits 28 - caf-20170518-22:45:58-cal-alex.fits 29 - caf-20170518-22:47:25-cal-alex.fits 30 - caf-20170518-22:48:51-cal-alex.fits 31 - caf-20170518-22:50:17-cal-alex.fits 32 - caf-20170518-22:51:43-cal-alex.fits 33 - caf-20170518-22:53:09-cal-alex.fits 34 - caf-20170518-22:54:36-cal-alex.fits 35 - caf-20170518-22:56:02-cal-alex.fits 36 - caf-20170518-22:57:28-cal-alex.fits 37 - caf-20170518-22:58:55-cal-alex.fits 38 - caf-20170518-23:00:21-cal-alex.fits 39 - caf-20170518-23:01:47-cal-alex.fits 40 - caf-20170518-23:03:13-cal-alex.fits 41 - caf-20170518-23:04:39-cal-alex.fits 42 - caf-20170518-23:06:05-cal-alex.fits 43 - caf-20170518-23:07:31-cal-alex.fits 44 - caf-20170518-23:08:58-cal-alex.fits 45 - caf-20170518-23:10:24-cal-alex.fits 46 - caf-20170518-23:11:50-cal-alex.fits 47 - caf-20170518-23:13:16-cal-alex.fits 48 - caf-20170518-23:14:42-cal-alex.fits 49 - caf-20170518-23:16:09-cal-alex.fits 50 - caf-20170518-23:17:35-cal-alex.fits 51 - caf-20170518-23:19:01-cal-alex.fits 52 - caf-20170518-23:20:27-cal-alex.fits 53 - caf-20170518-23:21:54-cal-alex.fits 54 - caf-20170518-23:23:20-cal-alex.fits 55 - caf-20170518-23:24:46-cal-alex.fits 56 - caf-20170518-23:26:12-cal-alex.fits 57 - caf-20170518-23:27:39-cal-alex.fits 58 - caf-20170518-23:29:05-cal-alex.fits 59 - caf-20170518-23:30:31-cal-alex.fits 60 - caf-20170518-23:31:57-cal-alex.fits 61 - caf-20170518-23:33:24-cal-alex.fits 62 - caf-20170518-23:34:50-cal-alex.fits 63 - caf-20170518-23:36:16-cal-alex.fits 64 - caf-20170518-23:37:42-cal-alex.fits 65 - caf-20170518-23:39:08-cal-alex.fits 66 - caf-20170518-23:40:35-cal-alex.fits 67 - caf-20170518-23:42:01-cal-alex.fits 68 - caf-20170518-23:43:27-cal-alex.fits 69 - caf-20170518-23:44:53-cal-alex.fits 70 - caf-20170518-23:46:19-cal-alex.fits 71 - caf-20170518-23:47:46-cal-alex.fits 72 - caf-20170518-23:49:12-cal-alex.fits 73 - caf-20170518-23:50:38-cal-alex.fits 74 - caf-20170518-23:52:05-cal-alex.fits 75 - caf-20170518-23:53:31-cal-alex.fits 76 - caf-20170518-23:54:57-cal-alex.fits 77 - caf-20170518-23:57:05-cal-alex.fits 78 - caf-20170518-23:58:31-cal-alex.fits 79 - caf-20170518-23:59:57-cal-alex.fits 80 - caf-20170519-00:01:23-cal-alex.fits 81 - caf-20170519-00:02:50-cal-alex.fits 82 - caf-20170519-00:04:16-cal-alex.fits 83 - caf-20170519-00:05:42-cal-alex.fits 84 - caf-20170519-00:07:09-cal-alex.fits 85 - caf-20170519-00:08:34-cal-alex.fits 86 - caf-20170519-00:10:01-cal-alex.fits 87 - caf-20170519-00:11:27-cal-alex.fits 88 - caf-20170519-00:12:53-cal-alex.fits 89 - caf-20170519-00:14:19-cal-alex.fits 90 - caf-20170519-00:15:46-cal-alex.fits 91 - caf-20170519-00:17:12-cal-alex.fits 92 - caf-20170519-00:18:38-cal-alex.fits 93 - caf-20170519-00:20:04-cal-alex.fits 94 - caf-20170519-00:21:30-cal-alex.fits 95 - caf-20170519-00:22:57-cal-alex.fits 96 - caf-20170519-00:24:23-cal-alex.fits 97 - caf-20170519-00:25:50-cal-alex.fits 98 - caf-20170519-00:27:17-cal-alex.fits 99 - caf-20170519-00:28:43-cal-alex.fits 100 - caf-20170519-00:30:09-cal-alex.fits 101 - caf-20170519-00:31:35-cal-alex.fits 102 - caf-20170519-00:33:02-cal-alex.fits 103 - caf-20170519-00:34:28-cal-alex.fits 104 - caf-20170519-00:35:55-cal-alex.fits 105 - caf-20170519-00:37:21-cal-alex.fits 106 - caf-20170519-00:38:47-cal-alex.fits 107 - caf-20170519-00:40:13-cal-alex.fits 108 - caf-20170519-00:41:40-cal-alex.fits 109 - caf-20170519-00:43:06-cal-alex.fits 110 - caf-20170519-00:44:32-cal-alex.fits 111 - caf-20170519-00:45:58-cal-alex.fits 112 - caf-20170519-00:47:24-cal-alex.fits 113 - caf-20170519-00:48:50-cal-alex.fits 114 - caf-20170519-00:50:16-cal-alex.fits 115 - caf-20170519-00:51:43-cal-alex.fits 116 - caf-20170519-00:53:09-cal-alex.fits 117 - caf-20170519-00:54:35-cal-alex.fits 118 - caf-20170519-00:56:01-cal-alex.fits 119 - caf-20170519-00:57:27-cal-alex.fits 120 - caf-20170519-00:58:53-cal-alex.fits 121 - caf-20170519-01:00:20-cal-alex.fits 122 - caf-20170519-01:01:46-cal-alex.fits 123 - caf-20170519-01:03:12-cal-alex.fits 124 - caf-20170519-01:04:38-cal-alex.fits 125 - caf-20170519-01:06:04-cal-alex.fits 126 - caf-20170519-01:07:30-cal-alex.fits 127 - caf-20170519-01:08:57-cal-alex.fits 128 - caf-20170519-01:10:23-cal-alex.fits 129 - caf-20170519-01:11:49-cal-alex.fits 130 - caf-20170519-01:13:16-cal-alex.fits 131 - caf-20170519-01:14:42-cal-alex.fits 132 - caf-20170519-01:16:08-cal-alex.fits 133 - caf-20170519-01:17:34-cal-alex.fits 134 - caf-20170519-01:19:45-cal-alex.fits 135 - caf-20170519-01:21:11-cal-alex.fits 136 - caf-20170519-01:22:38-cal-alex.fits 137 - caf-20170519-01:24:04-cal-alex.fits 138 - caf-20170519-01:25:30-cal-alex.fits 139 - caf-20170519-01:26:55-cal-alex.fits 140 - caf-20170519-01:28:22-cal-alex.fits 141 - caf-20170519-01:29:48-cal-alex.fits 142 - caf-20170519-01:31:14-cal-alex.fits 143 - caf-20170519-01:32:40-cal-alex.fits 144 - caf-20170519-01:34:07-cal-alex.fits 145 - caf-20170519-01:35:33-cal-alex.fits 146 - caf-20170519-01:36:59-cal-alex.fits 147 - caf-20170519-01:38:25-cal-alex.fits 148 - caf-20170519-01:39:52-cal-alex.fits 149 - caf-20170519-01:41:18-cal-alex.fits 150 - caf-20170519-01:42:44-cal-alex.fits 151 - caf-20170519-01:44:10-cal-alex.fits 152 - caf-20170519-01:45:36-cal-alex.fits 153 - caf-20170519-01:47:02-cal-alex.fits 154 - caf-20170519-01:48:28-cal-alex.fits 155 - caf-20170519-01:49:55-cal-alex.fits 156 - caf-20170519-01:51:21-cal-alex.fits 157 - caf-20170519-01:52:47-cal-alex.fits 158 - caf-20170519-01:54:13-cal-alex.fits 159 - caf-20170519-01:55:39-cal-alex.fits 160 - caf-20170519-01:57:06-cal-alex.fits 161 - caf-20170519-01:58:32-cal-alex.fits 162 - caf-20170519-01:59:58-cal-alex.fits 163 - caf-20170519-02:01:24-cal-alex.fits 164 - caf-20170519-02:02:50-cal-alex.fits 165 - caf-20170519-02:04:16-cal-alex.fits 166 - caf-20170519-02:05:43-cal-alex.fits 167 - caf-20170519-02:07:09-cal-alex.fits 168 - caf-20170519-02:08:35-cal-alex.fits 169 - caf-20170519-02:10:01-cal-alex.fits 170 - caf-20170519-02:11:27-cal-alex.fits 171 - caf-20170519-02:12:53-cal-alex.fits 172 - caf-20170519-02:14:20-cal-alex.fits 173 - caf-20170519-02:15:46-cal-alex.fits 174 - caf-20170519-02:17:12-cal-alex.fits 175 - caf-20170519-02:18:38-cal-alex.fits 176 - caf-20170519-02:20:05-cal-alex.fits 177 - caf-20170519-02:21:31-cal-alex.fits 178 - caf-20170519-02:22:57-cal-alex.fits 179 - caf-20170519-02:24:23-cal-alex.fits 180 - caf-20170519-02:25:50-cal-alex.fits 181 - caf-20170519-02:27:16-cal-alex.fits 182 - caf-20170519-02:28:42-cal-alex.fits 183 - caf-20170519-02:30:08-cal-alex.fits 184 - caf-20170519-02:31:34-cal-alex.fits 185 - caf-20170519-02:33:00-cal-alex.fits 186 - caf-20170519-02:34:26-cal-alex.fits 187replace-keyword: 188 - IMAGETYP: science 189 190--- 191 192# erroneous wrong-flat-imaging: they are actually science exposures 193night: 170601_t2_CAFOS 194enabled: True 195files: 196 - caf-20170601-22:54:57-cal-pelm.fits 197 - caf-20170601-23:06:07-cal-pelm.fits 198 - caf-20170601-23:16:42-cal-pelm.fits 199 - caf-20170601-23:29:19-cal-pelm.fits 200 - caf-20170601-23:30:24-cal-pelm.fits 201 - caf-20170601-23:31:28-cal-pelm.fits 202 - caf-20170601-23:38:56-cal-pelm.fits 203 - caf-20170601-23:49:59-cal-pelm.fits 204 - caf-20170602-00:00:43-cal-pelm.fits 205 - caf-20170602-00:12:12-cal-pelm.fits 206 - caf-20170602-00:13:17-cal-pelm.fits 207 - caf-20170602-00:14:22-cal-pelm.fits 208 - caf-20170602-00:19:25-cal-pelm.fits 209 - caf-20170602-00:30:01-cal-pelm.fits 210 - caf-20170602-00:40:41-cal-pelm.fits 211 - caf-20170602-00:54:30-cal-pelm.fits 212 - caf-20170602-00:55:34-cal-pelm.fits 213 - caf-20170602-01:01:14-cal-pelm.fits 214 - caf-20170602-01:13:10-cal-pelm.fits 215 - caf-20170602-01:24:38-cal-pelm.fits 216 - caf-20170602-01:36:43-cal-pelm.fits 217 - caf-20170602-01:39:11-cal-pelm.fits 218 - caf-20170602-01:40:24-cal-pelm.fits 219 - caf-20170602-01:43:45-cal-pelm.fits 220 - caf-20170602-01:55:03-cal-pelm.fits 221 - caf-20170602-02:06:16-cal-pelm.fits 222 - caf-20170602-02:17:44-cal-pelm.fits 223 - caf-20170602-02:18:49-cal-pelm.fits 224 - caf-20170602-02:19:54-cal-pelm.fits 225 - caf-20170602-02:22:30-cal-pelm.fits 226 - caf-20170602-02:34:59-cal-pelm.fits 227 - caf-20170602-02:47:55-cal-pelm.fits 228 - caf-20170602-03:02:56-cal-pelm.fits 229 - caf-20170602-03:04:00-cal-pelm.fits 230 - caf-20170602-03:05:05-cal-pelm.fits 231replace-keyword: 232 - IMAGETYP: science
Ignoring the initial comment lines (starting by
#), here we have created two blocks, separated by---(the YAML block separator). Important: the separator must not appear before the first block nor after the last block. Within each block, the following arguments must be provided:night: observing night. Note that wildcards can not be used here, although the same night label can appear in different blocks.enabled: this key indicates that the files included in this block are going to be fixed. Setting this key to False allows to disable a particular block without the need of removing it from this file.files: is the list of files to be fixed within the specified night. Although the list of files can be provided by given the name of each file in separate line, preceded by an indented-symbol, wildcards are also valid here.replace-keyword: list ofkeyword: new-value(one entry in each line). For this case, only the keywordsIMAGETYPis going to be set toscience. This small change will modify the next image classification of the involved images.
Once we have inserted the problematic images within this file, we can repeat the classification of these files. Since the images correspond to two different nights, it is necessary to repeat the classification in the corresponding nights (forcing the reduction with
--force):$ filabres -rs initialize -n 170518* --force ... ... $ filabres -rs initialize -n 170601* --force ... ...
Finally, it is possible to double check that the 210 images do not longer appear as classified as
wrong-flat-imaging:$ filabres -lc wrong-flat-imaging -k exptime -k quant500 -k quant975 -k object \ --filter "800 < k[quant500] < 50000" Total: 0 files
In fact, the 175 files from
170518appear now in the list of images classified asscience-imaging:$ filabres -lc science-imaging -n 170518* file 1 /Volumes/NicoPassport/CAHA/CAFOS2017/170518_t2_CAFOS/caf-20170518-22:21:06-cal-alex.fits 2 /Volumes/NicoPassport/CAHA/CAFOS2017/170518_t2_CAFOS/caf-20170518-22:22:33-cal-alex.fits 3 /Volumes/NicoPassport/CAHA/CAFOS2017/170518_t2_CAFOS/caf-20170518-22:23:59-cal-alex.fits ... ... 173 /Volumes/NicoPassport/CAHA/CAFOS2017/170518_t2_CAFOS/caf-20170519-02:31:34-cal-alex.fits 174 /Volumes/NicoPassport/CAHA/CAFOS2017/170518_t2_CAFOS/caf-20170519-02:33:00-cal-alex.fits 175 /Volumes/NicoPassport/CAHA/CAFOS2017/170518_t2_CAFOS/caf-20170519-02:34:26-cal-alex.fits Total: 175 filesThe same is true for the 35 images from
170601:$ filabres -lc science-imaging -n 170601* file 1 /Volumes/NicoPassport/CAHA/CAFOS2017/170601_t2_CAFOS/caf-20170601-20:37:01-sci-bomd.fits 2 /Volumes/NicoPassport/CAHA/CAFOS2017/170601_t2_CAFOS/caf-20170601-20:45:40-sci-bomd.fits 3 /Volumes/NicoPassport/CAHA/CAFOS2017/170601_t2_CAFOS/caf-20170601-20:50:37-sci-bomd.fits ... ... 36 /Volumes/NicoPassport/CAHA/CAFOS2017/170601_t2_CAFOS/caf-20170602-03:02:56-cal-pelm.fits 37 /Volumes/NicoPassport/CAHA/CAFOS2017/170601_t2_CAFOS/caf-20170602-03:04:00-cal-pelm.fits 38 /Volumes/NicoPassport/CAHA/CAFOS2017/170601_t2_CAFOS/caf-20170602-03:05:05-cal-pelm.fits Total: 38 files(Note that this last night contains 38 files and not 35: 3 science images do not belong to the set of files erroneously flagged as
wrong-flat-imagingin the initial classification.)
Wrong science-imaging
These are the science-imaging images that passed the requirements set in
configuration_cafos.yaml but not the requirmentx:
$ filabres -lc wrong-science-imaging
file
1 /Volumes/NicoPassport/CAHA/CAFOS2017/170502_t2_CAFOS/caf-20170503-00:58:59-sci-alex.fits
2 /Volumes/NicoPassport/CAHA/CAFOS2017/170526_t2_CAFOS/caf-20170527-03:57:08-sci-boeh.fits
3 /Volumes/NicoPassport/CAHA/CAFOS2017/170526_t2_CAFOS/caf-20170527-04:04:16-cal-boeh.fits
...
...
32 /Volumes/NicoPassport/CAHA/CAFOS2017/170528_t2_CAFOS/caf-20170528-16:37:50-cal-boeh.fits
33 /Volumes/NicoPassport/CAHA/CAFOS2017/170627_t2_CAFOS/caf-20170628-01:15:04-sci-mirl.fits
34 /Volumes/NicoPassport/CAHA/CAFOS2017/171217_t2_CAFOS/caf-20171217-17:38:26-sci-bard.fits
Total: 34 files
To understand why these images have been classified in this category, we can display additional keywords:
$ filabres -lc wrong-science-imaging -k object -k imagetyp -k exptime -k quant500 -k quant975
OBJECT IMAGETYP EXPTIME QUANT500 QUANT975 file
1 HAT-P-12b B science 0.0 698.0 727.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170502_t2_CAFOS/caf-20170503-00:58:59-sci-alex.fits
2 [IMG]RXCJ1913+7403 science 120.0 65531.0 65531.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170526_t2_CAFOS/caf-20170527-03:57:08-sci-boeh.fits
3 BIAS science 0.0 674.0 690.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170526_t2_CAFOS/caf-20170527-04:04:16-cal-boeh.fits
4 BIAS science 0.0 674.0 690.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170526_t2_CAFOS/caf-20170527-04:05:27-cal-boeh.fits
5 BIAS science 0.0 674.0 690.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170526_t2_CAFOS/caf-20170527-04:06:38-cal-boeh.fits
6 BIAS science 0.0 674.0 690.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170526_t2_CAFOS/caf-20170527-04:07:50-cal-boeh.fits
7 BIAS science 0.0 674.0 690.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170526_t2_CAFOS/caf-20170527-04:09:02-cal-boeh.fits
8 BIAS science 0.0 674.0 690.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170526_t2_CAFOS/caf-20170527-04:10:15-cal-boeh.fits
9 BIAS science 0.0 674.0 691.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170526_t2_CAFOS/caf-20170527-04:11:27-cal-boeh.fits
10 BIAS science 0.0 674.0 690.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170526_t2_CAFOS/caf-20170527-04:12:39-cal-boeh.fits
11 BIAS science 0.0 675.0 691.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170526_t2_CAFOS/caf-20170527-04:13:51-cal-boeh.fits
12 BIAS science 0.0 675.0 691.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170526_t2_CAFOS/caf-20170527-04:15:02-cal-boeh.fits
13 BIAS science 0.0 672.0 688.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170527_t2_CAFOS/caf-20170527-16:34:04-cal-boeh.fits
14 BIAS science 0.0 667.0 684.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170527_t2_CAFOS/caf-20170527-16:35:15-cal-boeh.fits
15 BIAS science 0.0 668.0 685.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170527_t2_CAFOS/caf-20170527-16:36:27-cal-boeh.fits
16 BIAS science 0.0 668.0 685.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170527_t2_CAFOS/caf-20170527-16:37:38-cal-boeh.fits
17 BIAS science 0.0 668.0 685.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170527_t2_CAFOS/caf-20170527-16:38:50-cal-boeh.fits
18 BIAS science 0.0 668.0 685.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170527_t2_CAFOS/caf-20170527-16:40:01-cal-boeh.fits
19 BIAS science 0.0 668.0 685.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170527_t2_CAFOS/caf-20170527-16:41:13-cal-boeh.fits
20 BIAS science 0.0 668.0 685.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170527_t2_CAFOS/caf-20170527-16:42:24-cal-boeh.fits
21 BIAS science 0.0 668.0 685.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170527_t2_CAFOS/caf-20170527-16:43:36-cal-boeh.fits
22 BIAS science 0.0 668.0 685.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170527_t2_CAFOS/caf-20170527-16:44:47-cal-boeh.fits
23 BIAS science 0.0 667.0 683.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170528_t2_CAFOS/caf-20170528-16:27:05-cal-boeh.fits
24 BIAS science 0.0 667.0 685.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170528_t2_CAFOS/caf-20170528-16:28:16-cal-boeh.fits
25 BIAS science 0.0 667.0 684.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170528_t2_CAFOS/caf-20170528-16:29:28-cal-boeh.fits
26 BIAS science 0.0 667.0 684.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170528_t2_CAFOS/caf-20170528-16:30:40-cal-boeh.fits
27 BIAS science 0.0 667.0 684.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170528_t2_CAFOS/caf-20170528-16:31:51-cal-boeh.fits
28 BIAS science 0.0 667.0 684.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170528_t2_CAFOS/caf-20170528-16:33:03-cal-boeh.fits
29 BIAS science 0.0 667.0 684.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170528_t2_CAFOS/caf-20170528-16:34:15-cal-boeh.fits
30 BIAS science 0.0 667.0 684.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170528_t2_CAFOS/caf-20170528-16:35:27-cal-boeh.fits
31 BIAS science 0.0 668.0 685.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170528_t2_CAFOS/caf-20170528-16:36:39-cal-boeh.fits
32 BIAS science 0.0 667.0 684.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170528_t2_CAFOS/caf-20170528-16:37:50-cal-boeh.fits
33 IRAS18061-2505 I science 900.0 33228.0 61843.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170627_t2_CAFOS/caf-20170628-01:15:04-sci-mirl.fits
34 HJ339 I science 120.0 65531.0 65531.0 /Volumes/NicoPassport/CAHA/CAFOS2017/171217_t2_CAFOS/caf-20171217-17:38:26-sci-bard.fits
Total: 34 files
Although a few of these images are saturated (just looking at QUANT500 and
QUANT975), most of them are actually bias frames (with EXPTIME=0)
wrongly acquired as science images.
It is possible to fix this error by using, again, the file
image_header_corrections.yaml.
For the particular problem of the wrong science images that are actually bias exposures, the new contents of this file are (only the last part of the file is displayed):
233
234---
235
236# erroneous science-imaging: it is actually a bias image
237night: 170502_t2_CAFOS
238enabled: True
239files:
240 - caf-20170503-00:58:59-sci-alex.fits
241replace-keyword:
242 - IMAGETYP: bias
243 - OBJECT: bias
244
245---
246
247# erroneous science-imaging: they are actually bias images
248night: 170526_t2_CAFOS
249enabled: True
250files:
251 - caf-20170527-04:04:16-cal-boeh.fits
252 - caf-20170527-04:05:27-cal-boeh.fits
253 - caf-20170527-04:06:38-cal-boeh.fits
254 - caf-20170527-04:07:50-cal-boeh.fits
255 - caf-20170527-04:09:02-cal-boeh.fits
256 - caf-20170527-04:10:15-cal-boeh.fits
257 - caf-20170527-04:11:27-cal-boeh.fits
258 - caf-20170527-04:12:39-cal-boeh.fits
259 - caf-20170527-04:13:51-cal-boeh.fits
260 - caf-20170527-04:15:02-cal-boeh.fits
261replace-keyword:
262 - IMAGETYP: bias
263 - OBJECT: bias
264
265---
266
267# erroneous science-imaging: they are actually bias images
268night: 170527_t2_CAFOS
269enabled: True
270files:
271 - caf-20170527-16:34:04-cal-boeh.fits
272 - caf-20170527-16:35:15-cal-boeh.fits
273 - caf-20170527-16:36:27-cal-boeh.fits
274 - caf-20170527-16:37:38-cal-boeh.fits
275 - caf-20170527-16:38:50-cal-boeh.fits
276 - caf-20170527-16:40:01-cal-boeh.fits
277 - caf-20170527-16:41:13-cal-boeh.fits
278 - caf-20170527-16:42:24-cal-boeh.fits
279 - caf-20170527-16:43:36-cal-boeh.fits
280 - caf-20170527-16:44:47-cal-boeh.fits
281replace-keyword:
282 - IMAGETYP: bias
283 - OBJECT: bias
284
285---
286
287# erroneous science-imaging: they are actually bias images
288night: 170528_t2_CAFOS
289enabled: True
290files:
291 - caf-20170528-16:27:05-cal-boeh.fits
292 - caf-20170528-16:28:16-cal-boeh.fits
293 - caf-20170528-16:29:28-cal-boeh.fits
294 - caf-20170528-16:30:40-cal-boeh.fits
295 - caf-20170528-16:31:51-cal-boeh.fits
296 - caf-20170528-16:33:03-cal-boeh.fits
297 - caf-20170528-16:34:15-cal-boeh.fits
298 - caf-20170528-16:35:27-cal-boeh.fits
299 - caf-20170528-16:36:39-cal-boeh.fits
300 - caf-20170528-16:37:50-cal-boeh.fits
301replace-keyword:
302 - IMAGETYP: bias
303 - OBJECT: bias
Remember that the YAML separator --- must not appear before the first block
nor after the last block.
Note that, for this particular problem with the wrong-science-imaging
files, four new blocks have been created, one for each night. The parameter
replace-keyword is defined with a list of keyword: new-value (one entry
in each line), being the list constituted by the two keywords IMAGETYP and
OBJECT. Both of them are going to be set to bias. This small change in
these keywords will modify the next image classification of the involved
images.
The list of files to be included in each of those four blocks have been
generated using (note the change of night -n <night>):
$ filabres -lc wrong-science-imaging --filter "k[quant975] < 800" -n 170502* -lm basic
- caf-20170503-00:58:59-sci-alex.fits
Total: 1 files
$ filabres -lc wrong-science-imaging --filter "k[quant975] < 800" -n 170526* -lm basic
- caf-20170527-04:04:16-cal-boeh.fits
- caf-20170527-04:05:27-cal-boeh.fits
- caf-20170527-04:06:38-cal-boeh.fits
- caf-20170527-04:07:50-cal-boeh.fits
- caf-20170527-04:09:02-cal-boeh.fits
- caf-20170527-04:10:15-cal-boeh.fits
- caf-20170527-04:11:27-cal-boeh.fits
- caf-20170527-04:12:39-cal-boeh.fits
- caf-20170527-04:13:51-cal-boeh.fits
- caf-20170527-04:15:02-cal-boeh.fits
Total: 10 files
$ filabres -lc wrong-science-imaging --filter "k[quant975] < 800" -n 170527* -lm basic
- caf-20170527-16:34:04-cal-boeh.fits
- caf-20170527-16:35:15-cal-boeh.fits
- caf-20170527-16:36:27-cal-boeh.fits
- caf-20170527-16:37:38-cal-boeh.fits
- caf-20170527-16:38:50-cal-boeh.fits
- caf-20170527-16:40:01-cal-boeh.fits
- caf-20170527-16:41:13-cal-boeh.fits
- caf-20170527-16:42:24-cal-boeh.fits
- caf-20170527-16:43:36-cal-boeh.fits
- caf-20170527-16:44:47-cal-boeh.fits
Total: 10 files
$ filabres -lc wrong-science-imaging --filter "k[quant975] < 800" -n 170528* -lm basic
- caf-20170528-16:27:05-cal-boeh.fits
- caf-20170528-16:28:16-cal-boeh.fits
- caf-20170528-16:29:28-cal-boeh.fits
- caf-20170528-16:30:40-cal-boeh.fits
- caf-20170528-16:31:51-cal-boeh.fits
- caf-20170528-16:33:03-cal-boeh.fits
- caf-20170528-16:34:15-cal-boeh.fits
- caf-20170528-16:35:27-cal-boeh.fits
- caf-20170528-16:36:39-cal-boeh.fits
- caf-20170528-16:37:50-cal-boeh.fits
Total: 10 files
Since the images affected by the erroneous keywords belong to four different
nights, namely 170502, 170526, 170527 and 170528, we are going
to repeat the full classification of all the images within only these four
nights (forcing the reduction with --force):
$ filabres -rs initialize -n 170502* --force
* Number of nights found: 1
* Working with night 170502_t2_CAFOS (1/1) ---> 408 FITS files
...
...
* program STOP
$ filabres -rs initialize -n 17052[678]* --force
* Number of nights found: 3
* Working with night 170526_t2_CAFOS (1/3) ---> 52 FITS files
...
...
* program STOP
Initially 826 images were classified as bias frames. Now, after the inclusion of the new 31 images, this number has increased to 857:
$ filabres -lc bias
...
...
Total: 857 files
In parallel, the number of wrong-science-imaging frames has decreased to
only 3 files (note the use of -pi argument to display the images on the
screen; here we are sorting by the median QUANT500):
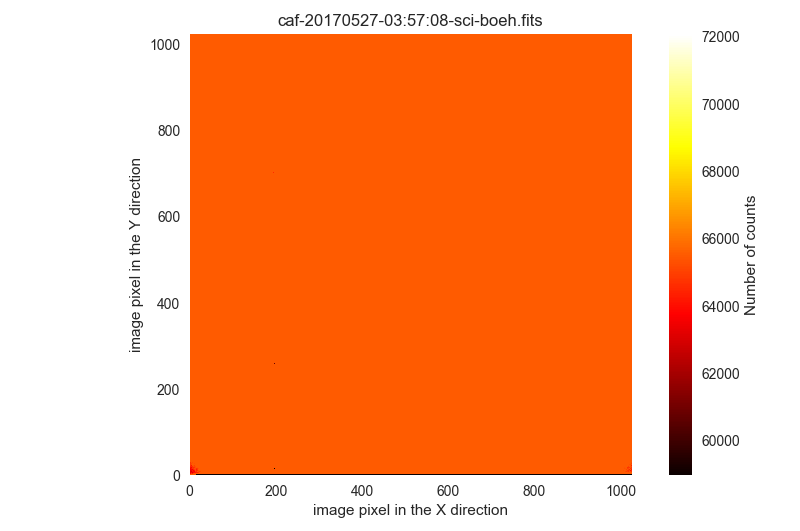
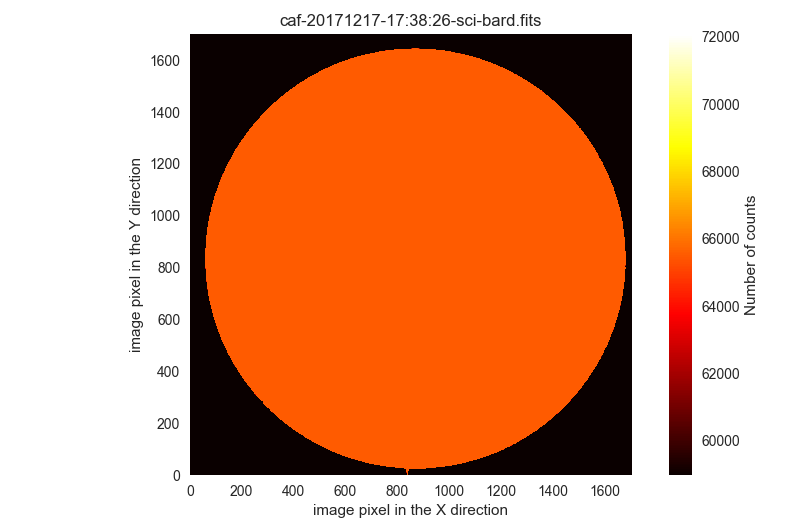
$ filabres -lc wrong-science-imaging -k object -k imagetyp -k exptime -ks quant500 -k quant975 -pi
2 IRAS18061-2505 I science 900.0 61843.0 33228.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170627_t2_CAFOS/caf-20170628-01:15:04-sci-mirl.fits
1 [IMG]RXCJ1913+7403 science 120.0 65531.0 65531.0 /Volumes/NicoPassport/CAHA/CAFOS2017/170526_t2_CAFOS/caf-20170527-03:57:08-sci-boeh.fits
3 HJ339 I science 120.0 65531.0 65531.0 /Volumes/NicoPassport/CAHA/CAFOS2017/171217_t2_CAFOS/caf-20171217-17:38:26-sci-bard.fits
Total: 3 files
The first image in this list is a scientific exposure with many saturated stars. We are going to ignore it:
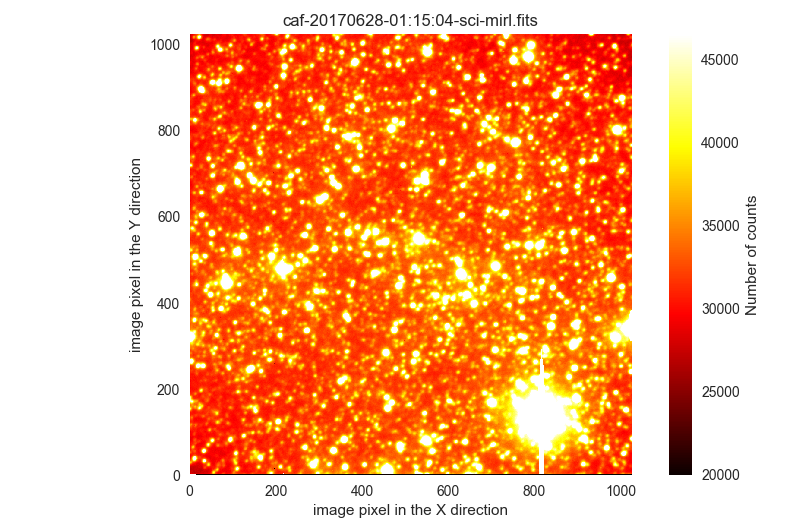
The last two images are clearly saturated and useless.