Reduction of science-imaging frames
Warning
The following documentation has been prepared executing the commands in a terminal running the bash shell. If you are using the Z shell (that at the time of writing is the default in mac OS) it is important to remember that the wildcards employed in some filabres parameters need to be provided enclosed in double quotes.
Note
The reduction of science images is carried out individually, i.e., the
scientific images are not combined within any time span prior to the
reduction process (in fact, the value of maxtimespan_hours is set to
zero in the file configuration_cafos.yaml).
There are many individual images classified as science-imaging:
$ filabres -lc science-imaging
...
...
Total: 4931 files
Only in the first night we have 48 images:
$ filabres -lc science-imaging -n 170225*
file
1 /Volumes/NicoPassport/CAHA/CAFOS2017/170225_t2_CAFOS/caf-20170225-18:59:12-sci-krek.fits
2 /Volumes/NicoPassport/CAHA/CAFOS2017/170225_t2_CAFOS/caf-20170225-19:01:30-sci-krek.fits
3 /Volumes/NicoPassport/CAHA/CAFOS2017/170225_t2_CAFOS/caf-20170225-19:04:18-sci-krek.fits
...
...
46 /Volumes/NicoPassport/CAHA/CAFOS2017/170225_t2_CAFOS/caf-20170225-22:17:06-sci-krek.fits
47 /Volumes/NicoPassport/CAHA/CAFOS2017/170225_t2_CAFOS/caf-20170225-22:18:58-sci-krek.fits
48 /Volumes/NicoPassport/CAHA/CAFOS2017/170225_t2_CAFOS/caf-20170225-22:20:27-sci-krek.fits
Total: 48 files
Considering the large number of images, it is advisable to start the reduction
of these scientific images by constraining the process to a subset of images.
In this sense, we can choose between the different options shown below. Note
that in all the following examples we are using enhanced verbosity (argument
-v/--verbose) and an interactive execution (argument
-i/--interactive; this is going to stop the reduction to show some
intermediate plots and results).
The different possibilities are (do not try these examples yet! their purpose here is simply to illustrate the different approaches):
Reduce a single
science-imagingimage using the argument-n/--nightfor the night and the argument--filenamefollowed by the explicit name of the original FITS file (without the file path). For example, the first file of the first night is reduced executing:$ filabres -rs science-imaging -v -i -n 170225* --filename caf-20170225-18:59:12-sci-krek.fits
Here we have specified the reduction step
-rs science-imaging, the use of enhanced verbosity-v, the interactive execution of the program-i, the observing night-n 170225*, and the specific file to be reduced--filename caf-20170225-18:59:12-sci-krek.fits.Reduce all the
science-imagingfiles within a given night. In this case just omit--filename <file>. Only the night must be specified:$ filabres -rs science-imaging -v -i -n 170225*
In this case you will probably prefer to execute the reduction of the files without
-i.Reduce all the
science-imagingfiles of all nights: if a specific night is not given, filabres will try to reduce all the images of this type, independently of the observing night:$ filabres -rs science-imaging -v -i
In this case you will also prefer to execute the reduction of the files without
-i(and also without-v; relevant information concerning the reduction of each individual file is conveniently stored).
In the next section we are describing in detail the first scenario, i.e. the
reduction of a single science-imaging frame. The description is also valid
for the other to cases, where the reduction is extended to a large number of
files.
Reduction of a single science-imaging frame
Warning
Note that the astrometric calibration is performed using GAIA data downloaded from the internet on real time while executing filabres. This means that a live internet connection is required for the code to work properly.
We illustrate this case using the first science-imaging file of the first
night. As previously mentioned, the reduction of this particular image starts
by executing:
$ filabres -rs science-imaging -v -i -n 170225* --filename caf-20170225-18:59:12-sci-krek.fits
Warning
Do not get alarmed by the verbosity of the output generated by the reduction of this type of images. Here we are describing that output little by little.
Note that you do not have to type anything of what is shown in this section:
all the contents displayed are output generated by filabres, including
the execution of external programs (displayed after the $ symbol) which
is automatically handled by the program.
The initial output shows basic information concerning the configuration files employed by filabres:
* instrument: cafos
* datadir: /Volumes/NicoPassport/CAHA/CAFOS2017
* ignored_images_file: ignored_images.yaml
* image_header_corrections_file: image_header_corrections.yaml
* forced_classifications_file: forced_classifications.yaml
* Loading instrument configuration
* Number of nights found: 1
* List of nights: ['170225_t2_CAFOS']
Subdirectory science-imaging not found. Creating it!
A new subdirectory, called science-imaging, has been created under the
current directory. This new subdirectory will host a tree of subdirectories
(corresponding to different observing nights) where the reduction of the images
will take place and the associated files will be stored.
* Working with night 170225_t2_CAFOS (1/1)
Reading file ./lists/170225_t2_CAFOS/imagedb_cafos.json
Number of science-imaging images found: 1
Subdirectory science-imaging/170225_t2_CAFOS not found. Creating it!
Since we have specified a single image corresponding to an individual file,
filabres locates that image. In addition, since that image is the first one
to be reduced corresponding to the night 170225_t2_CAFOS, a subdirectory
with that name is created under the subdirectory science-imaging.
Note
The information displayed in the terminal while executing the reduction
steps is also stored in an auxiliary file called reduction.log, which is
placed in the same directory where other auxiliary files, generated during
the astrometric calibration, are also stored (as explained below).
Basic reduction of caf-20170225-18:59:12-sci-krek.fits
Results database set to science-imaging/170225_t2_CAFOS/filabres_db_cafos_science-imaging.json
The basic information concerning the reduction of the image will be stored in a
database called filabres_db_cafos_science-imaging.json. Note that contrary
to what is done with the reduced calibrations, where all the information is
stored in a single database filabres_db_cafos_<calibration>.json (where
<calibration> is bias or flat-imaging), in the case of the science
images, that information is separately stored in independent files
filabres_db_science-imaging.json located within the subdirectory reserved
for each observing night under the science-imaging subdirectory tree. In
addition to this, within the same subdirectory tree, a specific subdirectory is
also created for each reduced image, where filabres stores additional
auxiliary files created during the data reduction. This will be explained with
more detail below.
---
-> Working with file caf-20170225-18:59:12-sci-krek.fits (1/1) [Night 1/1]
-> Input file name is......: /Volumes/NicoPassport/CAHA/CAFOS2017/170225_t2_CAFOS/caf-20170225-18:59:12-sci-krek.fits
-> Output file name will be: science-imaging/170225_t2_CAFOS/science-imaging_caf-20170225-18:59:12-sci-krek_red.fits
-> Reduction starts at.....: 2020-03-25 10:23:23.597982
The absolute path to the input file name is shown, as well as the relative path
to the main output file, which basename is identical to the input file, adding
the previx science-imaging_ and the suffix _red (the latter prior to
the extension .fits).
Basic reduction of the image
The basic reduction of the image includes the bias subtraction and the flatfielding.
Calibration database set to filabres_db_cafos_bias.json
-> looking for calibration bias with signature SITE#1d_15__1650__1650__[251,221:1900,1870]__1__1
-> mjdobsarray.......: [57808.89853]
-> looking for mjdobs: 57809.7911
-> nearest value is..: 57808.89853
-> delta_mjd (days)..: -0.8925700000036159
The master bias is retrieved by looking into the database
filabres_db_cafos_bias.json, and locating all the reduced master bias
frames with the expected signature (just one in this example). When several
master bias frames are available (with the required signature), the one closest
to the observation time of the scientific image is employed.
Calibration database set to filabres_db_cafos_flat-imaging.json
-> looking for calibration flat-imaging with signature SITE#1d_15__1650__1650__[251,221:1900,1870]__1__1__GRISM-11__FILT- 9__FREE__0
-> mjdobsarray.......: [57808.85161 57810.2688 ]
-> looking for mjdobs: 57809.7911
-> nearest value is..: 57810.26880
-> delta_mjd (days)..: 0.4776999999958207
The same action is performed with the flatfield. The master flatfield is
retrieved from the database filabres_db_cafos_flat-imaging.json.
Astrometric calibration: description of the method
The astrometric calibration is performed in two steps:
Using Astrometry.net utilities: this provides an initial astrometric solution, making use of a gnomic projection with SIP (Simple Imaging Polynomial) distortions:
CTYPE1='RA---TAN-SIP'andCTYPE2='DEC--TAN-SIP'.The required binaries are:
build-astrometry-index: computes a suitable index file (containing hash codes of typically sets of four stars) that facilitates the alignment of the requested image. Note that filabres does not use the pre-computed index files provided by Astrometry.net, but uses index files especially suited for each region of the sky covered by the science images. These files are built from GAIA data downloaded from the internet while executing the code.Within each night, a database called
central_pointings.jsonis created with the regions of the sky covered by the images already reduced for that observing night. This avoids the need to download GAIA data for images that correspond to close pointings by reusing already download data.solve-field: determines the astrometric calibration using the index file previously computed.
The initial astrometric calibration provides typical errors of the order of the seeing, although we have checked that these errors are larger at the image borders in a systematic way, probably because the distortion is determined using second-order polynomials, which is not good enough. For that reason, this astrometric calibration is refined by using the AstrOmatic.net tools.
Using AstrOmatic.net tools:
SExtractorandSCAMPare employed to detect the image sources and perform a refined astrometric calibration, using the TPV World Coordinate System to map the image distortions. The initial WCS solution provided by the Astrometry.net software allowsSCAMPto determine a much better WCS solution by setting the TPV polynomial degrees to 3, leading to typical errors within a fraction of a pixel. Again, GAIA data is retrieved from the internet to carry out this astrometric calibration.
In the next subsections we describe how these two software packages are employed by filabres.
Astrometric calibration with Astrometry.net tools
Astrometric calibration of science-imaging/170225_t2_CAFOS/science-imaging_caf-20170225-18:59:12-sci-krek_red.fits
Creating configuration file science-imaging/170225_t2_CAFOS/work/myastrometry.cfg
Central coordinates:
<SkyCoord (FK5: equinox=2017-02-25T18:59:12.000): (ra, dec) in deg
(58.824742, 53.26594)>
<SkyCoord (FK5: equinox=J2000.000): (ra, dec) in deg
(58.49578022, 53.21627399)>
A temporary work subdirectory is created under the subdirectory of the
observing night 170225_t2_CAFOS. This is the location where all the required
files will be placed in order to carry out the astrometric calibration.
The central coordinates of the field of view are read from the original FITS
keywords RA and DEC.
Downloading the GAIA data
Subdirectory science-imaging/170225_t2_CAFOS/index000001 not found. Creating it!
-> Creating science-imaging/170225_t2_CAFOS/index000001/gaialog.log
...
(several WARNING messages to be ignored)
...
The central coordinates of the image to be processed are not close to any
previous location of the sky for the considered night (in fact, this is the
firs image to be reduced within that night). The data we are going to download
from the GAIA catalogue will be stored in a particular subdirectory
index000001. Those data will be reused for the astrometric calibration of
other images obtained in the same sky region.
The file gaialog.log stores the GAIA query employed to download the data.
Here we are using the Table Access Protocol (TAP) developed by IVOA to
perform the data retrieval.
-> Gaia data: magnitude, nobjects: 30.000, 1602
Querying GAIA data: 1602 objects found
There is a limit of 2000 sources that can be retrieved in a single synchronous query to the GAIA data server. In this example, the number of sources, down to magnitude 30.0, is 1602, which is already below that limit of 2000. In more crowded fields, filabres will adjust the limiting magnitude in order to get the maximum number of sources, without exceeding 2000.
-> Applying proper motion correction...
...
(several WARNING messages to be ignored)
...
-> Saving science-imaging/170225_t2_CAFOS/index000001/GaiaDR2-query.fits
$ cp science-imaging/170225_t2_CAFOS/index000001/GaiaDR2-query.fits science-imaging/170225_t2_CAFOS/work/
The downloaded GAIA sources are corrected from proper motion. The corrected
catalogue is called GaiaDR2-query.fits (a binary table in FITS format). The
file is not only stored under index000001, but also
copied into the work subdirectory for its immediate use.
Generating reduced image science-imaging/170225_t2_CAFOS/work/xxx.fits (after bias subtraction and flatfielding)
A temporary version of the reduced scientific image (after bias subtraction and
flatfielding), named xxx.fits, has also been saved into the work
subdirectory.
Building the index file
At this point, filabres launches the execution of
build-astrometry-index within the work subdirectory, generating the
index file index-image.fits:
*** Using Astrometry.net tools ***
[Working in science-imaging/170225_t2_CAFOS/work]
$ build-astrometry-index -i GaiaDR2-query.fits -o index-image.fits -A ra -D dec -S phot_g_mean_mag -P 2 -E -I 1
The output generated by the previous command is large:
Reading GaiaDR2-query.fits...
Got 1602 stars
Sweep 1: 24 stars
Sweep 2: 24 stars
Sweep 3: 24 stars
Sweep 4: 24 stars
Sweep 5: 24 stars
Sweep 6: 23 stars
Sweep 7: 23 stars
Sweep 8: 23 stars
Sweep 9: 21 stars
Sweep 10: 21 stars
Total: 231 stars
Writing output...
First RA,Dec: 58.4315,53.2728
First x,y,z: 0.313067,0.50951,0.801491
Treetype: 0x10404
After kdtree_build:
kdtree:
type 0x10404
lr 0x7fc084409490
perm 0x7fc0844090f0
bb 0x0
nbb 0
split 0x7fc0844094d0
splitdim 0x0
dimbits 2
dimmask 0x3
splitmask 0xfffffffc
data 0x7fc084804600
free data 1
range [-1, 1] [-1, 1] [-1, 1]
scale 2.14748e+09
invscale 4.65661e-10
Ndata 231
Ndim 3
Nnodes 31
Nbottom 16
Ninterior 15
Nlevels 5
has_linear_lr 0
name stars
First data elements in tree: -929911,-1.19922e-116,-5.7116e+186
Reading star kdtree /tmp/tmp.skdt.MrLOgt ...
Will write to quad file /tmp/tmp.quad.Rm6hWZ and code file /tmp/tmp.code.lLGtFD
Nside=880. Nside^2=774400. Number of healpixes=9292800. Healpix side length ~ 3.99766 arcmin.
Star tree contains 231 objects.
Healpix radius 169.606 arcsec, quad scale 168 arcsec, total 340.982 arcsec
Scanning 231 input stars...
Will check 24 healpixes.
Pass 1 of 16.
Trying 24 healpixes.
.......................
Made 21 quads (out of 24 healpixes) this pass.
Made 21 quads so far.
Merging quads...
Pass 2 of 16.
Trying 24 healpixes.
.......................
Made 21 quads (out of 24 healpixes) this pass.
Made 42 quads so far.
Merging quads...
Pass 3 of 16.
Trying 24 healpixes.
.......................
Made 21 quads (out of 24 healpixes) this pass.
Made 63 quads so far.
Merging quads...
Pass 4 of 16.
Trying 24 healpixes.
.......................
Made 21 quads (out of 24 healpixes) this pass.
Made 84 quads so far.
Merging quads...
Pass 5 of 16.
Trying 24 healpixes.
.......................
Made 21 quads (out of 24 healpixes) this pass.
Made 105 quads so far.
Merging quads...
Pass 6 of 16.
Trying 24 healpixes.
.......................
Made 21 quads (out of 24 healpixes) this pass.
Made 126 quads so far.
Merging quads...
Pass 7 of 16.
Trying 24 healpixes.
.......................
Made 20 quads (out of 24 healpixes) this pass.
Made 146 quads so far.
Merging quads...
Pass 8 of 16.
Trying 24 healpixes.
.......................
Made 20 quads (out of 24 healpixes) this pass.
Made 166 quads so far.
Merging quads...
Pass 9 of 16.
Trying 24 healpixes.
.......................
Made 20 quads (out of 24 healpixes) this pass.
Made 186 quads so far.
Merging quads...
Pass 10 of 16.
Trying 24 healpixes.
.......................
Made 20 quads (out of 24 healpixes) this pass.
Made 206 quads so far.
Merging quads...
Pass 11 of 16.
Trying 24 healpixes.
.......................
Made 20 quads (out of 24 healpixes) this pass.
Made 226 quads so far.
Merging quads...
Pass 12 of 16.
Trying 24 healpixes.
.......................
Made 20 quads (out of 24 healpixes) this pass.
Made 246 quads so far.
Merging quads...
Pass 13 of 16.
Trying 24 healpixes.
.......................
Made 20 quads (out of 24 healpixes) this pass.
Made 266 quads so far.
Merging quads...
Pass 14 of 16.
Trying 24 healpixes.
.......................
Made 20 quads (out of 24 healpixes) this pass.
Made 286 quads so far.
Merging quads...
Pass 15 of 16.
Trying 24 healpixes.
.......................
Made 20 quads (out of 24 healpixes) this pass.
Made 306 quads so far.
Merging quads...
Pass 16 of 16.
Trying 24 healpixes.
.......................
Made 20 quads (out of 24 healpixes) this pass.
Made 326 quads so far.
Merging quads...
Loosening reuse maximum to 9...
Trying 58 healpixes.
.........................................................
Made 1 quads (out of 58 healpixes) this pass.
Loosening reuse maximum to 10...
Trying 57 healpixes.
........................................................
Made 1 quads (out of 57 healpixes) this pass.
Loosening reuse maximum to 11...
Trying 56 healpixes.
.......................................................
Made 1 quads (out of 56 healpixes) this pass.
Loosening reuse maximum to 12...
Trying 55 healpixes.
......................................................
Made 1 quads (out of 55 healpixes) this pass.
Loosening reuse maximum to 13...
Trying 54 healpixes.
.....................................................
Made 1 quads (out of 54 healpixes) this pass.
Loosening reuse maximum to 14...
Trying 53 healpixes.
....................................................
Made 1 quads (out of 53 healpixes) this pass.
Loosening reuse maximum to 15...
Trying 52 healpixes.
...................................................
Made 1 quads (out of 52 healpixes) this pass.
Loosening reuse maximum to 16...
Trying 51 healpixes.
..................................................
Made 1 quads (out of 51 healpixes) this pass.
Loosening reuse maximum to 17...
Trying 50 healpixes.
.................................................
Made 1 quads (out of 50 healpixes) this pass.
Loosening reuse maximum to 18...
Trying 49 healpixes.
................................................
Made 1 quads (out of 49 healpixes) this pass.
Loosening reuse maximum to 19...
Trying 48 healpixes.
...............................................
Made 0 quads (out of 48 healpixes) this pass.
Loosening reuse maximum to 20...
Trying 48 healpixes.
...............................................
Made 0 quads (out of 48 healpixes) this pass.
Writing quads...
Used 0.092124 s user, 0.00265 s system (0.094774 s total), 0.094853 s wall time since last check
Done.
codetree: building KD tree for /tmp/tmp.code.lLGtFD
will write KD tree file /tmp/tmp.ckdt.qcwc4Z
Reading codes...
Read 336 codes.
Building tree...
Done
Writing code KD tree to /tmp/tmp.ckdt.qcwc4Z...
Unpermute-stars...
Unpermuting stars from /tmp/tmp.skdt.MrLOgt and /tmp/tmp.quad.Rm6hWZ to /tmp/tmp.skdt2.qInvwK and /tmp/tmp.quad2.c9YLzG
Reading star tree from /tmp/tmp.skdt.MrLOgt ...
Reading quadfile from /tmp/tmp.quad.Rm6hWZ ...
Writing quadfile to /tmp/tmp.quad2.c9YLzG ...
Writing quads...
...............................................................................
Writing star kdtree to /tmp/tmp.skdt2.qInvwK ...
Permuting tag-along table...
Unpermute-quads...
Unpermuting quads from /tmp/tmp.quad2.c9YLzG and /tmp/tmp.ckdt.qcwc4Z to /tmp/tmp.quad3.b42npc and /tmp/tmp.ckdt2.1PzOO1
Reading code tree from /tmp/tmp.ckdt.qcwc4Z ...
Reading quads from /tmp/tmp.quad2.c9YLzG ...
Writing quads to /tmp/tmp.quad3.b42npc ...
Writing code kdtree to /tmp/tmp.ckdt2.1PzOO1 ...
Merging /tmp/tmp.quad3.b42npc and /tmp/tmp.ckdt2.1PzOO1 and /tmp/tmp.skdt2.qInvwK to index-image.fits
Reading code tree from /tmp/tmp.ckdt2.1PzOO1 ...
Ok.
Reading star tree from /tmp/tmp.skdt2.qInvwK ...
Ok.
Reading quads from /tmp/tmp.quad3.b42npc ...
Ok.
Solving the field
Next, the field corresponding to the file xxx.fits (the reduced image)
is solved using the index file just computed:
[Working in science-imaging/170225_t2_CAFOS/work]
$ solve-field -p --config myastrometry.cfg --overwrite --ra 58.495780218141974 --dec 53.21627398668612 --radius 0.13333333333333333 xxx.fits
Reading input file 1 of 1: "xxx.fits"...
Extracting sources...
simplexy: found 185 sources.
Solving...
Reading file "./xxx.axy"...
Only searching for solutions within 0.133333 degrees of RA,Dec (58.4958,53.2163)
Field 1 did not solve (index index-image.fits, field objects 1-10).
mo field stars:
star 10; field_xy 616.2,1298.1, field_orig 616.2,1298.1
star 1; field_xy 760.8,823.6, field_orig 760.8,823.6
star 2; field_xy 773.8,1227.7, field_orig 773.8,1227.7
star 3; field_xy 524.6,1148.6, field_orig 524.6,1148.6
log-odds ratio 182.725 (2.27127e+79), 63 match, 2 conflict, 87 distractors, 162 index.
RA,Dec = (58.497,53.2251), pixel scale 0.525226 arcsec/pix.
Hit/miss: Hit/miss: ++-+--+-+-----c+--+---+++--++-+--+++-+---++++----++++---++------+++--+-+---+--++++---+--+---+-++--+-
Field 1: solved with index index-image.fits.
Field 1 solved: writing to file ./xxx.solved to indicate this.
Field: xxx.fits
Field center: (RA,Dec) = (58.497161, 53.225216) deg.
Field center: (RA H:M:S, Dec D:M:S) = (03:53:59.319, +53:13:30.779).
Field size: 14.3725 x 14.3903 arcminutes
Field rotation angle: up is 86.6417 degrees E of N
Field parity: pos
Creating new FITS file "./xxx.new"...
The previous command finds the sources, generating a binary FITS table called
xxx.axy. Immediately after that, the field is solved using the location of
those sources. The file xxx.new is a first version of the reduced image
containing an astrometric calibration.
Before leaving the astrometric calibration with the Astrometry.net tools,
filabres determines if any of the sources found in the image xxx.fits
is saturated. If this is the case, the saturated objects can be removed from
the file xxx.axy, which allows to solve again for a new astrometric
solution. Note that the new execution of solve-field uses as input the
updated xxx.axy file instead of the original xxx.fits image.
Checking file: science-imaging/170225_t2_CAFOS/work/xxx.axy
Number of saturated objects found: 1/185
Saturated object: (1167.2056, 576.06335, 64436.547, 84.54297)
File: science-imaging/170225_t2_CAFOS/work/xxx.axy updated
[Working in science-imaging/170225_t2_CAFOS/work]
$ solve-field -p --config myastrometry.cfg --continue --width 1650 --height 1650 --x-column X --y-column Y --sort-column FLUX --ra 58.495780218141974 --dec 53.21627398668612 --radius 0.13333333333333333 xxx.axy
Reading input file 1 of 1: "xxx.axy"...
Reading sort column "FLUX"
Sorting sort column
mmapping input file
Copying table header.
Writing row 0
Done
Solving...
Reading file "./xxx.axy"...
Only searching for solutions within 0.133333 degrees of RA,Dec (58.4958,53.2163)
Field 1 did not solve (index index-image.fits, field objects 1-10).
mo field stars:
star 10; field_xy 616.2,1298.1, field_orig 616.2,1298.1
star 0; field_xy 760.8,823.6, field_orig 760.8,823.6
star 1; field_xy 773.8,1227.7, field_orig 773.8,1227.7
star 2; field_xy 524.6,1148.6, field_orig 524.6,1148.6
log-odds ratio 177.04 (7.72096e+76), 62 match, 2 conflict, 87 distractors, 162 index.
RA,Dec = (58.497,53.2251), pixel scale 0.525226 arcsec/pix.
Hit/miss: Hit/miss: ++-+--+-+-----c+--+---+-+--++-+--+++-++--++++----+++----++------+++--+-+---+--+-++---+-------++++-+-
Field 1: solved with index index-image.fits.
Field 1 solved: writing to file ./xxx.solved to indicate this.
Field: xxx.axy
Field center: (RA,Dec) = (58.497172, 53.225206) deg.
Field center: (RA H:M:S, Dec D:M:S) = (03:53:59.321, +53:13:30.742).
Field size: 14.3712 x 14.3898 arcminutes
Field rotation angle: up is 86.638 degrees E of N
Field parity: pos
The last execution of solve-field, which used xxx.axy as input, instead
of xxx.fits, does not automatically generate a final image xxx.new with
the astrometric calibration incorporated into the FITS header. Instead, only
the WCS solution is saved in the file xxx.wcs. For that reason, we have to
insert that WCS solution into the reduced image xxx.fits. Fortunately, the
utility new-wcs performs this tasks for us:
[Working in science-imaging/170225_t2_CAFOS/work]
$ new-wcs -i xxx.fits -w xxx.wcs -o xxx.new -d
WARNING: FITSFixedWarning: The WCS transformation has more axes (2) than the image it is associated with (0) [astropy.wcs.wcs]
Results of the initial astrometric calibration
The last output corresponding to the execution of Astrometry.net tools is a brief summary displaying the plate scales (arcsec/pix), the number of sources found, the mean error (in arcsec), and some of the outliers.
astrometry.net> pixel scales (arcsec/pix): [0.52343268 0.52341089]
astrometry-net> Number of targest found: 71
astrometry-net> Mean error (arcsec)....: 1.1879683209341652
-> outlier point #68, delta_r (arcsec): 5.620631094480526
-> outlier point #69, delta_r (arcsec): 8.548273785024582
-> outlier point #70, delta_r (arcsec): 8.712019148806954
-> outlier point #71, delta_r (arcsec): 10.140383487868942
In addition to the displayed numerical summary, several plots are created showing the result of the astrometric calibration obtained with Astrometry.net. Note that when running the program interactively, you can use all the options that are available in the navigation toolbar of the matplotlib windows, including pan, zoom, etc.
The first plot shows the error between the predicted location of the sources (using the derived astrometric solution) and the peak positions found in the image. Each object is labelled with a number, that increases with the total error, i.e. distance to the (0,0) point in the plot:
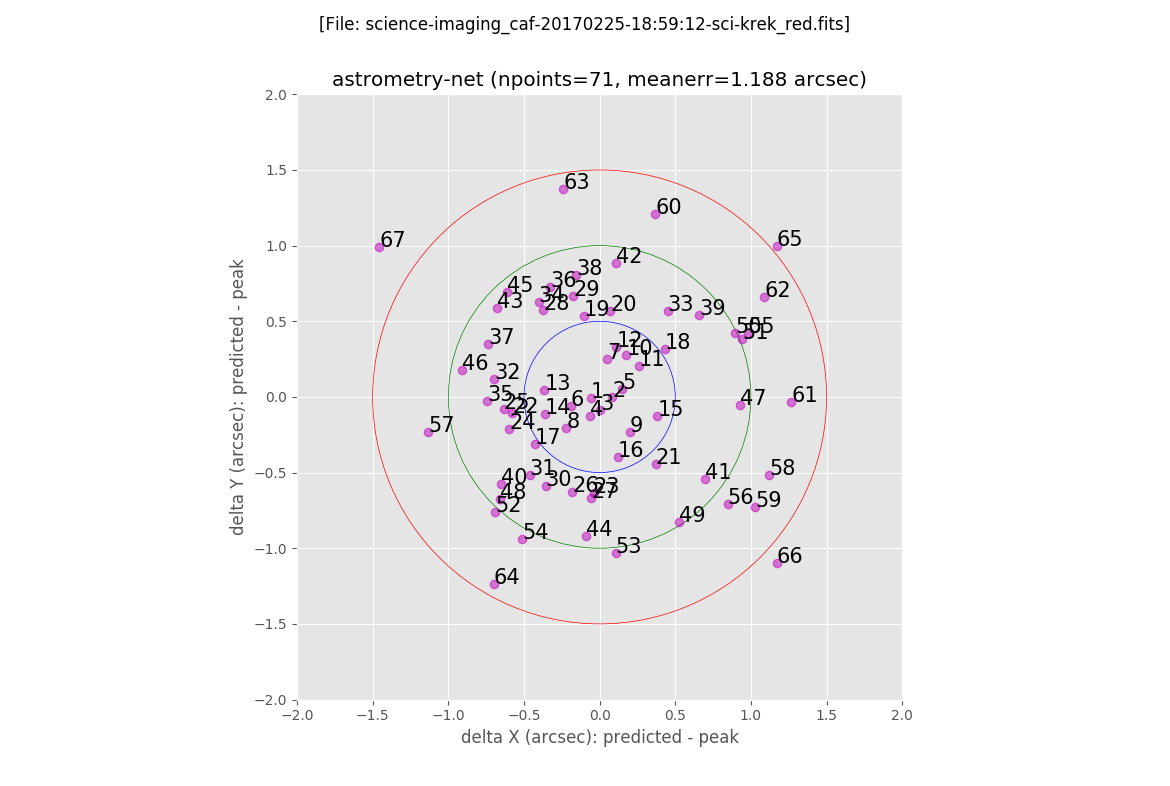
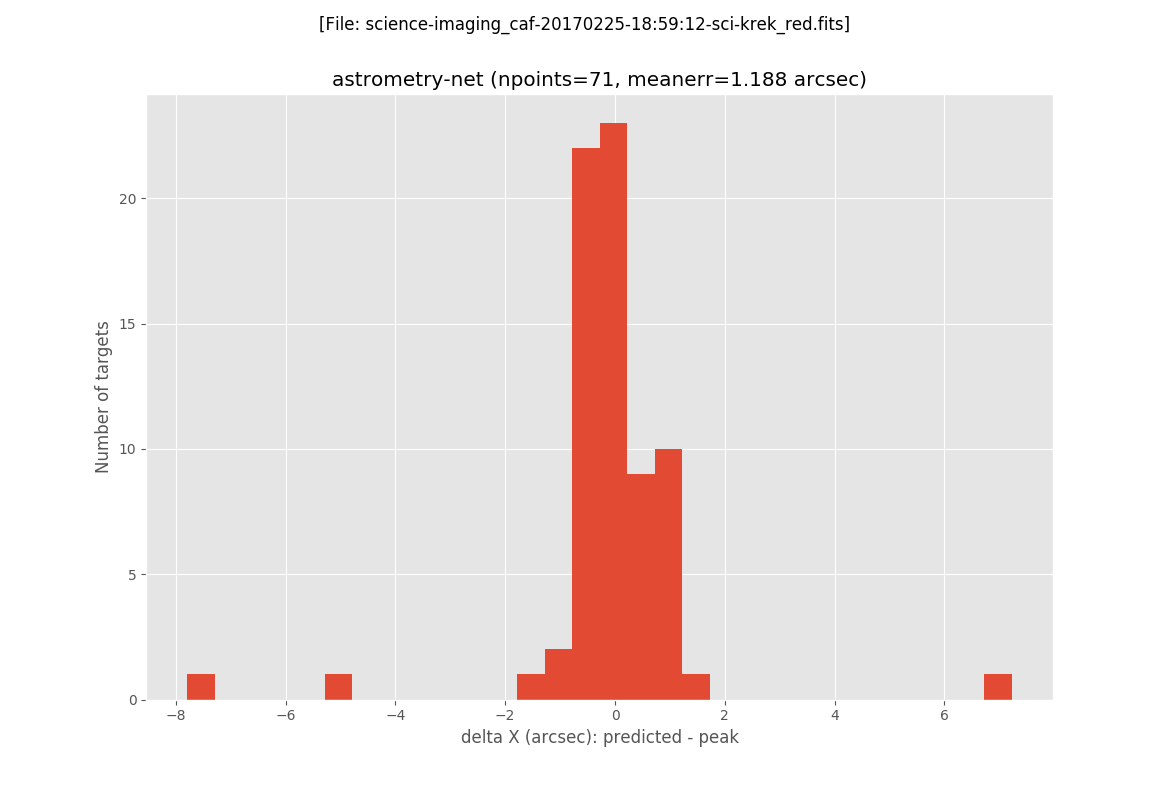
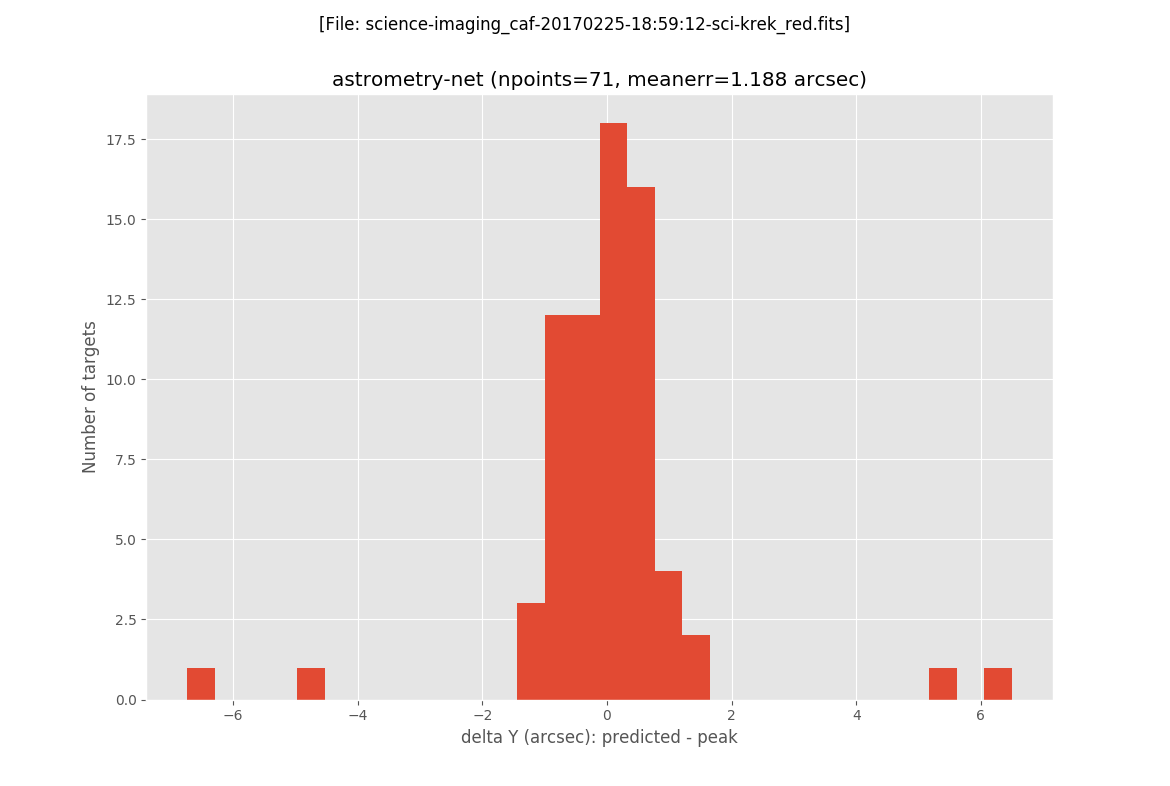
The next two plots are histograms with the errors in X and Y:
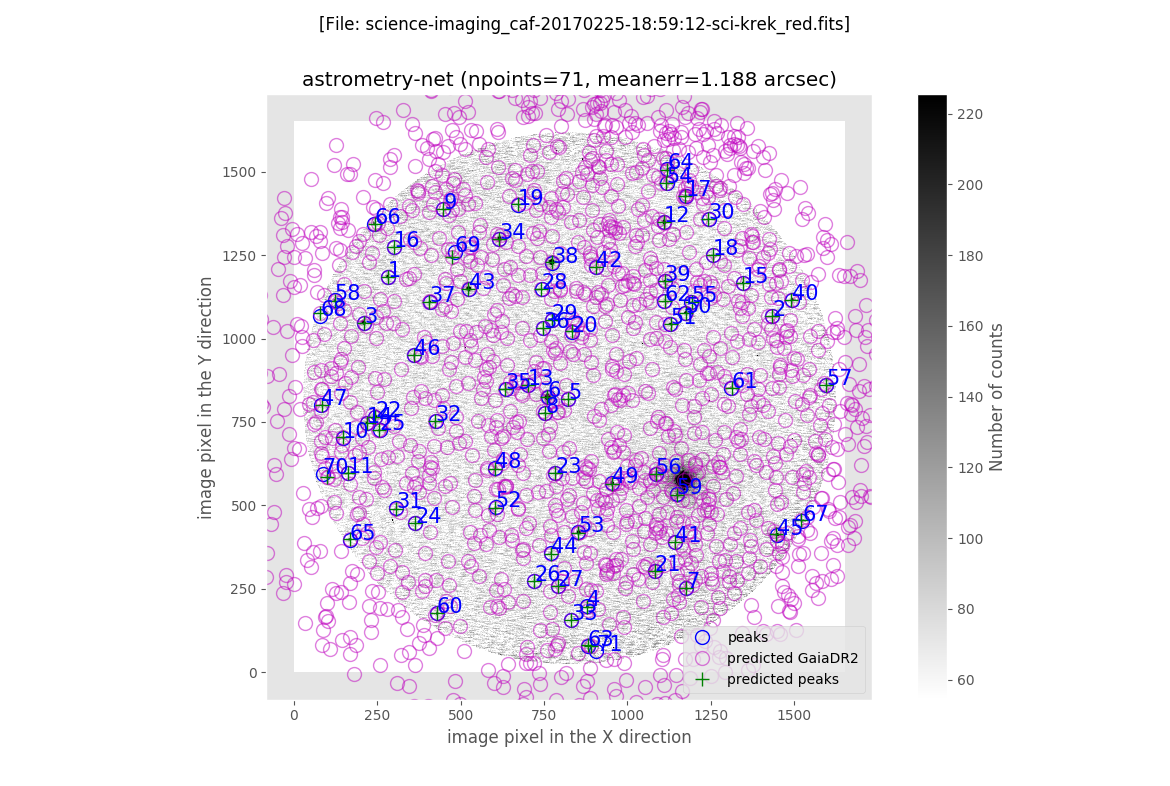
The final plot is an image of the scientific image: the blue circles represent the peak of the detected sources, the magenta crosses are the predicted location of all the GAIA sources in a slightly larger field of view (the one for wich the GAIA data was retrieved), and the green crosses indicate the predicted positions of the detected sources.
Astrometric calibration with AstrOmatic.net tools
The initial astrometric calibration obtained with the Astrometry.net tools is refined now using the AstrOmatic software.
Creating the configuration files
The first step is the generation of the configuration files required by
SExtractor and SCAMP.
*** Using AstrOmatic.net tools ***
Generating science-imaging/170225_t2_CAFOS/work/default.param
Generating science-imaging/170225_t2_CAFOS/work/config.sex
Generating science-imaging/170225_t2_CAFOS/work/config.scamp
Running ``SExtractor``
Next, SExtractor is executed in order to build the catalog of objects in
the reduced image, which will be called xxx.ldac.
[Working in science-imaging/170225_t2_CAFOS/work]
$ sex xxx.new -c config.sex -CATALOG_NAME xxx.ldac
The output of the execution of SExtractor is also shown:
>
----- SExtractor 2.25.0 started on 2020-03-20 at 20:42:49 with 1 thread
> Setting catalog parameters
> Initializing catalog
> Looking for xxx.new
----- Measuring from: xxx.new
"IC342 g'" / no ext. header / 1650x1650 / 32 bits (floats)
Detection+Measurement image: > Setting up background maps
> Setting up background map at line: 64
> Setting up background map at line: 128
> Setting up background map at line: 192
> Setting up background map at line: 256
> Setting up background map at line: 320
> Setting up background map at line: 384
> Setting up background map at line: 448
> Setting up background map at line: 512
> Setting up background map at line: 576
> Setting up background map at line: 640
> Setting up background map at line: 704
> Setting up background map at line: 768
> Setting up background map at line: 832
> Setting up background map at line: 896
> Setting up background map at line: 960
> Setting up background map at line: 1024
> Setting up background map at line: 1088
> Setting up background map at line: 1152
> Setting up background map at line: 1216
> Setting up background map at line: 1280
> Setting up background map at line: 1344
> Setting up background map at line: 1408
> Setting up background map at line: 1472
> Setting up background map at line: 1536
> Setting up background map at line: 1600
> Filtering background map(s)
> Computing background d-map
> Computing background-noise d-map
(M+D) Background: 68.3404 RMS: 13.9395 / Threshold: 69.6973
> Scanning image
> Line: 25 Objects: 0 detected / 0 sextracted
> Line: 50 Objects: 7 detected / 0 sextracted
> Line: 75 Objects: 19 detected / 0 sextracted
> Line: 100 Objects: 20 detected / 0 sextracted
> Line: 125 Objects: 32 detected / 0 sextracted
> Line: 150 Objects: 32 detected / 0 sextracted
> Line: 175 Objects: 32 detected / 0 sextracted
> Line: 200 Objects: 33 detected / 0 sextracted
> Line: 225 Objects: 34 detected / 0 sextracted
> Line: 250 Objects: 35 detected / 0 sextracted
> Line: 275 Objects: 36 detected / 0 sextracted
> Line: 300 Objects: 38 detected / 0 sextracted
> Line: 325 Objects: 38 detected / 0 sextracted
> Line: 350 Objects: 38 detected / 0 sextracted
> Line: 375 Objects: 38 detected / 0 sextracted
> Line: 400 Objects: 38 detected / 0 sextracted
> Line: 425 Objects: 40 detected / 0 sextracted
> Line: 450 Objects: 41 detected / 0 sextracted
> Line: 475 Objects: 43 detected / 0 sextracted
> Line: 500 Objects: 43 detected / 0 sextracted
> Line: 525 Objects: 43 detected / 0 sextracted
> Line: 550 Objects: 44 detected / 0 sextracted
> Line: 575 Objects: 46 detected / 0 sextracted
> Line: 600 Objects: 46 detected / 0 sextracted
> Line: 625 Objects: 48 detected / 0 sextracted
> Line: 650 Objects: 48 detected / 0 sextracted
> Line: 675 Objects: 48 detected / 0 sextracted
> Line: 700 Objects: 48 detected / 0 sextracted
> Line: 725 Objects: 48 detected / 0 sextracted
> Line: 750 Objects: 49 detected / 0 sextracted
> Line: 775 Objects: 51 detected / 0 sextracted
> Line: 800 Objects: 52 detected / 0 sextracted
> Line: 825 Objects: 52 detected / 0 sextracted
> Line: 850 Objects: 53 detected / 0 sextracted
> Line: 875 Objects: 54 detected / 0 sextracted
> Line: 900 Objects: 56 detected / 0 sextracted
> Line: 925 Objects: 56 detected / 0 sextracted
> Line: 950 Objects: 60 detected / 0 sextracted
> Line: 975 Objects: 61 detected / 0 sextracted
> Line: 1000 Objects: 61 detected / 0 sextracted
> Line: 1025 Objects: 62 detected / 0 sextracted
> Line: 1034 Objects: 62 detected / 0 sextracted
> Line: 1050 Objects: 63 detected / 3 sextracted
> Line: 1075 Objects: 64 detected / 3 sextracted
> Line: 1100 Objects: 65 detected / 4 sextracted
> Line: 1125 Objects: 68 detected / 6 sextracted
> Line: 1150 Objects: 70 detected / 6 sextracted
> Line: 1175 Objects: 72 detected / 7 sextracted
> Line: 1200 Objects: 74 detected / 8 sextracted
> Line: 1225 Objects: 74 detected / 8 sextracted
> Line: 1250 Objects: 75 detected / 10 sextracted
> Line: 1275 Objects: 75 detected / 10 sextracted
> Line: 1300 Objects: 75 detected / 12 sextracted
> Line: 1325 Objects: 76 detected / 12 sextracted
> Line: 1350 Objects: 77 detected / 12 sextracted
> Line: 1375 Objects: 79 detected / 12 sextracted
> Line: 1400 Objects: 79 detected / 12 sextracted
> Line: 1425 Objects: 79 detected / 12 sextracted
> Line: 1450 Objects: 79 detected / 12 sextracted
> Line: 1475 Objects: 79 detected / 12 sextracted
> Line: 1500 Objects: 79 detected / 12 sextracted
> Line: 1525 Objects: 80 detected / 12 sextracted
> Line: 1550 Objects: 81 detected / 12 sextracted
> Line: 1575 Objects: 95 detected / 12 sextracted
> Line: 1600 Objects: 95 detected / 12 sextracted
> Line: 1625 Objects: 95 detected / 12 sextracted
> Line: 1650 Objects: 95 detected / 12 sextracted
Objects: detected 95 / sextracted 53
> Closing files
>
> All done (in 0.1 s: 14925.8 lines/s , 479.4 detections/s)
Running ``SCAMP``
Finally, SCAMP is executed to compute the refined astrometric calibration.
[Working in science-imaging/170225_t2_CAFOS/work]
$ scamp xxx.ldac -c config.scamp
The output of the execution of SCAMP is shown next:
> WARNING: FGROUP_RADIUS keyword unknown
> WARNING: Obsolete configuration; replacing cocat1.u-strasbg.fr with vizier.u-strasbg.fr
----- SCAMP 2.7.8 started on 2020-03-20 at 20:42:49 with 8 threads
----- 1 inputs:
xxx.ldac: "IC342 g' " no ext. header 1 set 50 detections
----- 50 detections loaded
----- 1 instrument found for astrometry:
Instrument A1 :
1 extensions
FILTER =
QRUNID =
----- 1 instrument found for photometry:
Instrument P1 :
FILTER =
----- 1 field group found:
Group 1: 1 field at 03:53:59.31 +53:13:30.8 with radius 10.18'
instruments epoch center coordinates radius scale
xxx.ldac A1 P1 2017.2 03:53:59.31 +53:13:30.8 10.18' 0.5234"
----- Reference catalogs:
Group 1: 13689 standards found in GAIA-DR2 (G band)
----- Astrometric matching:
Group 1: 13689 standards in GAIA-DR2 (band G)
instruments pos.angle scale cont. shift cont.
xxx.ldac A1 P1 +0.00 deg 0.5234" 0.697 +0.051" -0.30" 3.45
----- Astrometric clipping:
Group 1: 4/34 detections removed
----- Astrometric stats (internal) :
All detections | High S/N
dAXIS1 dAXIS2 chi2 ndets | dAXIS1 dAXIS2 chi2 ndets
Group 1: 0" 0" 0 29 0" 0" 0 10
----- Astrometric stats (external):
All detections | High S/N
dAXIS1 dAXIS2 chi2 nstars | dAXIS1 dAXIS2 chi2 nstars
Group 1: 0.0526" 0.0487" 6.1 29 0.0249" 0.0112" 10 10
----- Photometric clipping:
Group 1 / P1 : 0/29 detections removed
----- Photometric stats (internal):
All detections | High S/N
Instru mag RMS chi2 ndets | mag RMS chi2 ndets
Group 1: P1 0 0 29 0 0 10
----- Photometric stats (external):
All detections | High S/N
Instru mag RMS chi2 nstars | mag RMS chi2 nstars
Results of the new astrometric calibration
The next output is a brief summary displaying the plate scales (arcsec/pix), the number of sources found, the mean error (in arcsec), and some of the outliers:
-> file science-imaging/170225_t2_CAFOS/science-imaging_caf-20170225-18:59:12-sci-krek_red.fits created
astrometry> pixel scales (arcsec/pix): [0.52343268 0.52341089]
Reading science-imaging/170225_t2_CAFOS/work/full_1.cat
X_IMAGE is located in column #6
Y_IMAGE is located in column #7
CATALOG_NUMBER is located in column #2
Number of objects read: 50
Reading science-imaging/170225_t2_CAFOS/work/merged_1.cat
ALPHA_J2000 is located in column #4
DELTA_J2000 is located in column #5
astrometry-scamp> Number of targest found: 50
astrometry-scamp> Mean error (arcsec)....: 0.032813825746896653
-> outlier point #47, delta_r (arcsec): 0.15799448046263975
-> outlier point #48, delta_r (arcsec): 0.1599908735307402
-> outlier point #49, delta_r (arcsec): 0.18797255871764804
-> outlier point #50, delta_r (arcsec): 0.19884266054034697
In addition, filabres generates the same plots previously displayed when computing the astrometric solution with the Astrometry.net software. In this case it is clear the reduction of the error on the astrometric solution.
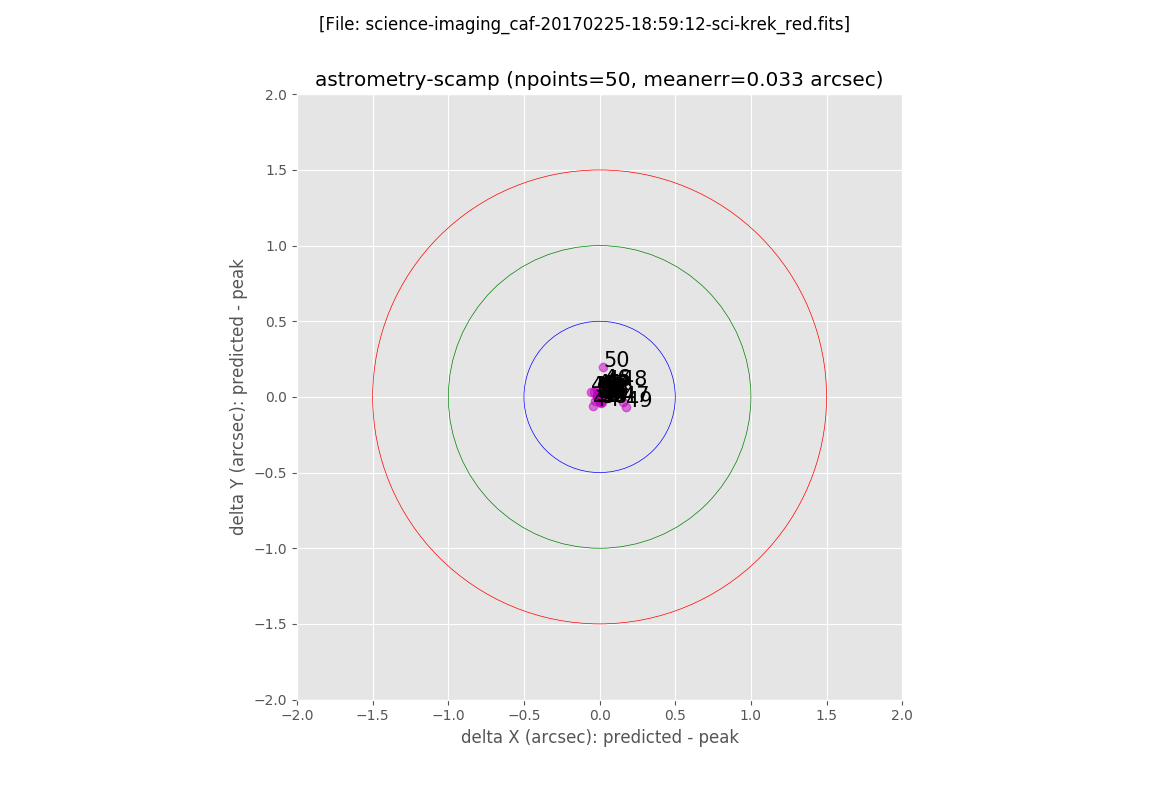
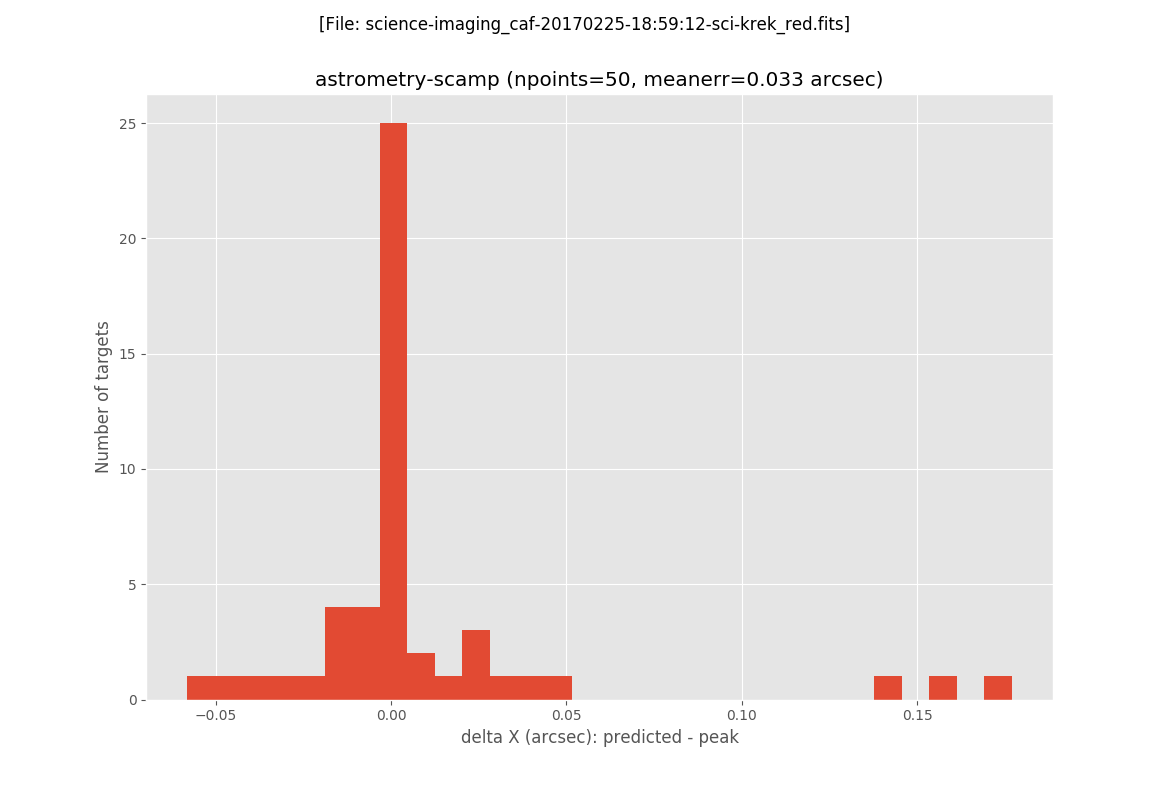
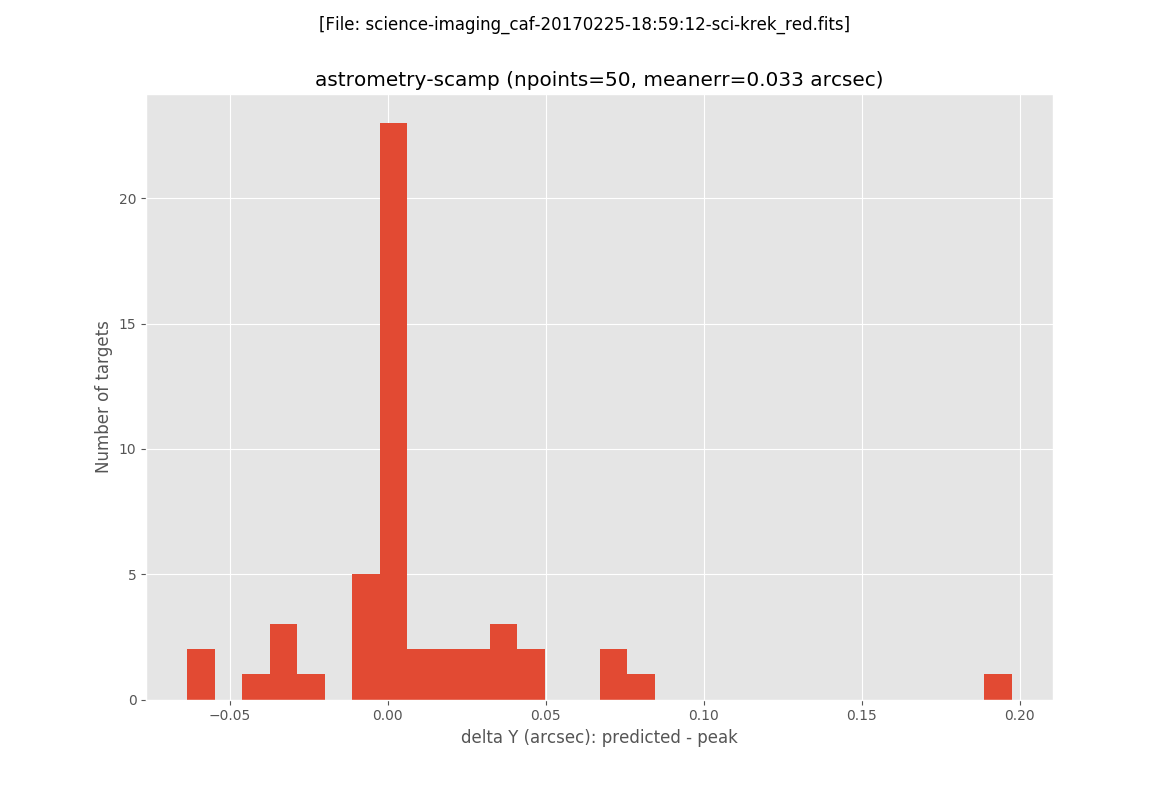
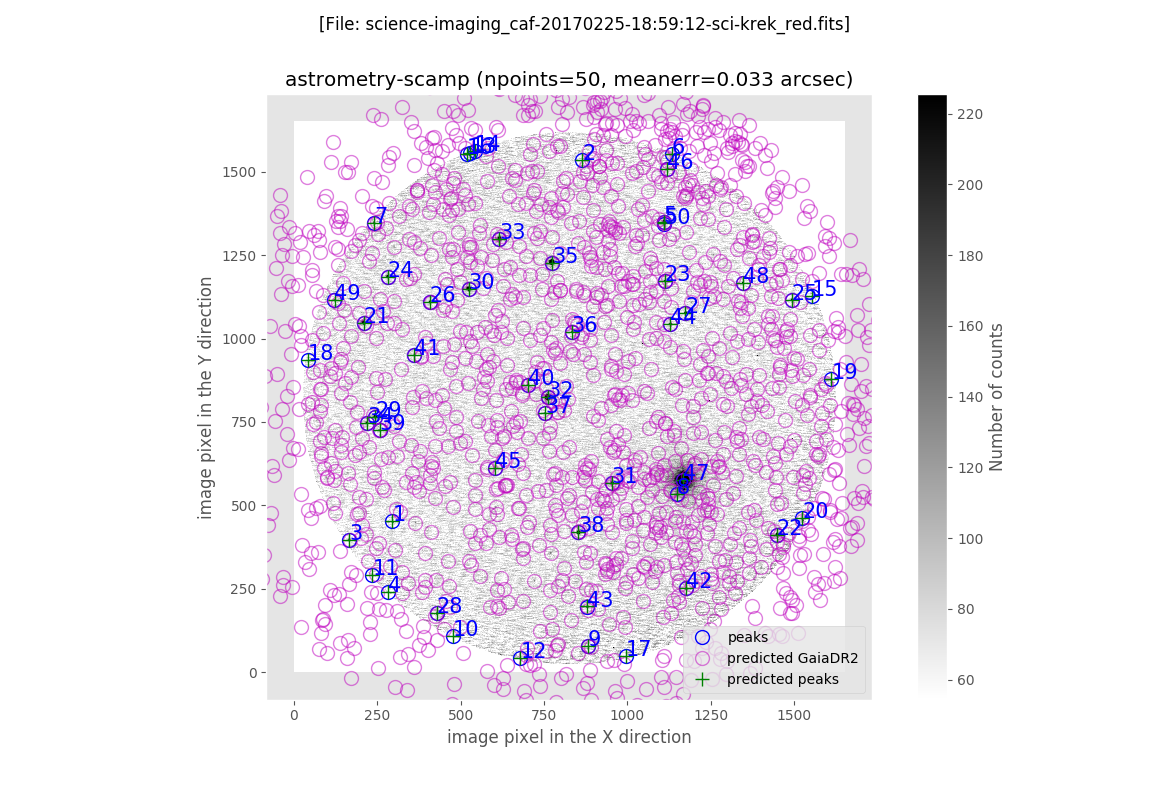
The final image after the execution of the AstrOmatic.net tools is then generated:
-> file science-imaging/170225_t2_CAFOS/science-imaging_caf-20170225-18:59:12-sci-krek_red.fits created
Storing the results
Filabres stores not only the reduced image, but also some auxiliary files
associated to the astrometric calibration. All that information is kept under
a subdirectory with the same name as the reduced image (without the .fits
extension):
Subdirectory science-imaging/170225_t2_CAFOS/science-imaging_caf-20170225-18:59:12-sci-krek_red not found. Creating it!
[Working in science-imaging/170225_t2_CAFOS/work]
$ cp astrometry-net.pdf ../science-imaging_caf-20170225-18:59:12-sci-krek_red/
[Working in science-imaging/170225_t2_CAFOS/work]
$ cp astrometry-scamp.pdf ../science-imaging_caf-20170225-18:59:12-sci-krek_red/
[Working in science-imaging/170225_t2_CAFOS/work]
$ cp xxx.new ../science-imaging_caf-20170225-18:59:12-sci-krek_red/
[Working in science-imaging/170225_t2_CAFOS/work]
$ cp full_1.cat ../science-imaging_caf-20170225-18:59:12-sci-krek_red/
[Working in science-imaging/170225_t2_CAFOS/work]
$ cp merged_1.cat ../science-imaging_caf-20170225-18:59:12-sci-krek_red/
-> Reduction ends at..,,...: 2020-03-25 10:24:06.429420
-> Elapsed time............: 0:00:42.831438
Saving reduction.log
$ mv science-imaging/170225_t2_CAFOS/reduction.log science-imaging/170225_t2_CAFOS/science-imaging_caf-20170225-18:59:12-sci-krek_red/
Press 'x' + <ENTER> to stop, or simply <ENTER> to continue...
Those files include the log file reduction.log (with all the relevant
information concerning the basic reduction and the output of the astrometric
calibration), the PDF version of the previously displayed plots, the reduced
image after the initial astrometric calibration with Astrometry.net (this file
is called xxx.new), and some catalogues generated by AstrOmatic.net.
Repeating the reduction
If you need to repeat the reduction of a particular image that has already been
reduced, remember to use the parameter --force. Otherwise the reduction is
skipped.
Reducing multiple science-imaging files
The previous sections have illustrated the reduction of a single
science-imaging frame. As previously mentioned, this process can be
trivially executed with all the similar images in a particular night by
avoiding the use of the --filename argument. In addition, it is also
convenient to exclude the use of -i/--interactive to prevent filabres
from pausing the reduction after finishing each scientific image:
$ filabres -rs science-imaging -v -n 170225*
or even to all nights by removing the -n/--night argument:
$ filabres -rs science-imaging -v
Checking the science-imaging reduction
To check the results of the science-imaging files, you can use the
-lr/--list_reduced argument, as previouly described for the calibration
images.
$ filabres -lr science-imaging
...
...
$ filabres -lr science-imaging -k all
...
...
It is interesting to check whether there has been any error when retrieveing the bias and flatfield calibrations, or during the astrometric calibration.
Problems with the bias calibration
$ filabres -lr science-imaging --filter "k[ierr_bias] != 0" -k ccdsec -k naxis1 -k naxis2
...
...
In these cases, no bias frame has been found with the requested signature. The median from the nearest bias image is subtracted instead (a constant value for the whole image).
Problems with the flat calibration
$ filabres -lr science-imaging --filter "k[ierr_flat] != 0" -k object -k ccdsec -k naxis1 -k naxis2
...
...
In these cases, no flat-imaging frame has been found with the requested signature. No flatfielding has been applied.
Problems with the astrometric calibration
$ filabres -lr science-imaging --filter "k[ierr_astr] != 0" -k astr1_meanerr -k astr2_meanerr
...
...
astr1_meanerr: mean error in the initial astrometric calibration with the Astrometry.net tools. IfNaN, the astrometric calibration has failed and only the basic reduction (bias and flatfielding) has been applied to the scientific image.astr2_meanerr: mean error in the refined astrometric calibration with AstrOmatic.net tools. IfNanbutastr1_meanerris notNan, the reduced scientific image at least contains the initial astrometric calibration performed with the Astrometry.net tools.
Removing invalid reduced science-imaging frames
To remove a particular reduced science-imaging result, it is important to
delete not only the actual FITS file, but also the associated files
stored during the data reduction, as well as the corresponding entry in the
database file filabres_db_cafos_science-imaging.json. Fortunately, all
these actions are performed automatically by filabres using the
--delete argument followed by the full path to the reduced file:
filabres --delete science-imaging/170225_t2_CAFOS/science-imaging_caf-20170225-18:59:12-sci-krek_red.fits
Image to be deleted science-imaging/170225_t2_CAFOS/science-imaging_caf-20170225-18:59:12-sci-krek_red.fits
-> Deleting entry in science-imaging/170225_t2_CAFOS/filabres_db_cafos_science-imaging.json
-> Updating science-imaging/170225_t2_CAFOS/filabres_db_cafos_science-imaging.json
-> Deleting file: science-imaging/170225_t2_CAFOS/science-imaging_caf-20170225-18:59:12-sci-krek_red.fits
-> Removing file: science-imaging/170225_t2_CAFOS/science-imaging_caf-20170225-18:59:12-sci-krek_red/astrometry-scamp.pdf
-> Removing file: science-imaging/170225_t2_CAFOS/science-imaging_caf-20170225-18:59:12-sci-krek_red/astrometry-net.pdf
-> Removing file: science-imaging/170225_t2_CAFOS/science-imaging_caf-20170225-18:59:12-sci-krek_red/merged_1.cat
-> Removing file: science-imaging/170225_t2_CAFOS/science-imaging_caf-20170225-18:59:12-sci-krek_red/full_1.cat
-> Removing file: science-imaging/170225_t2_CAFOS/science-imaging_caf-20170225-18:59:12-sci-krek_red/xxx.new
-> Removing file: science-imaging/170225_t2_CAFOS/science-imaging_caf-20170225-18:59:12-sci-krek_red/reduction.log
-> Removing subdirectory: science-imaging/170225_t2_CAFOS/science-imaging_caf-20170225-18:59:12-sci-krek_red
* program STOP
Skipping the astrometric calibration
It is possible to skip the astrometric calibration by using the argument
-na/--no_astrometry.